A Low-Density Protein Microarray Device for Detecting Three Antibodies Associated with Invasive Fungal Infections
Xiao Chen, Fang-qiu Li*, Chun-fang Ma, Li-ning Shi and Yu-an Hu
DOI10.21767/2471-8521.100019
Laboratory of Molecular Biology, Institute of Medical Laboratory Sciences, Jinling Hospital, School of Medicine, Nanjing University, Nanjing, 210002, PR China
- *Corresponding Author:
- Fang-qiu Li
Laboratory of Molecular Biology, Institute of Medical Laboratory Sciences, Jinling Hospital, 305 East Zhongshan Road, Nanjing 210002, P. R. China
Tel: +86-13801583645
E-mail: njlifq@163.com
Received Date: August 12, 2016; Accepted Date: September 29, 2016; Published Date: September 30, 2016
Citation: Chen X, Li F, Ma C. et al. A Low- Density Protein Microarray Device for Detecting Three Antibodies Associated with Invasive Fungal Infections. Med Mycol Open Access. 2016, 2:19. doi: 10.21767/2471-8521.100019
Copyright: © 2016 Li F, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Abstract
The frequency of invasive fungal infections has risen dramatically in recent years, and early and accurate diagnosis of these infections is important for appropriate use of antifungal agents. The value of serological tests is apparent, but they suffer from disadvantages, such as being time-consuming and their inability to produce simultaneous results. Protein microarrays are a convenient technology that can be used to perform high-throughput antibody detection and enable the monitoring of different antibodies simultaneously. We constructed a low-density microarray using purified recombinant enolase (Eno) fructose-bisphosphate aldolase (Fba1) from Candida and thioredoxin reductase (TR) from Aspergillus fumigatus as immobilized antigens. Following optimization of microarray construction and serum testing, we investigated the sensitivity, repeatability, and stability of the assay using sera from patients with invasive Candida and/or Aspergillus infections and control subjects. Sera from patients with invasive candidiasis (IC) were subjected to low-density microarray assay for detection of two antibodies from Candida and one antibody from A. fumigatus. Our results indicated a sensitivity and specificity of 67.6% and 96.5% for anti-Eno and 64.8% and 90.3% for anti- Fba1 antibodies, respectively. The combined detection of anti-Eno and anti- Fba1 returned a sensitivity of 81%, and in invasive aspergillosis (IA) patients, the sensitivity and specificity of detection were 71.4% and 98.2%, respectively, for the anti-TR antibody. The low-density protein microarray assay developed in this study was capable of detecting three antibodies against two fungi simultaneously, demonstrating its potential value in diagnosing IC and IA.
Keywords
Low-density protein microarray; Antibodies; Candida; Aspergillus
Introduction
Invasive fungal infections (IFIs) remain a major cause of mortality in those undergoing hematopoietic stem cell or solid-organ transplantation, chemotherapy for hematological malignancies, patients in intensive care units, and those infected with human immunodeficiency virus. Invasive candidiasis (IC) and invasive aspergillosis (IA) are associated with high mortality rates [1]. Candida spp. Are found in the environment in soil and water and frequently colonize human skin and mucosal membranes, with endogenous colonization responsible for most IC cases. Candidiasis covers infections that range from superficial mucosal (such as vagina) to systemic and potentially life-threatening diseases [2,3]. Aspergillus fumigatus is a saprophytic mold responsible for life-threatening, invasive pulmonary diseases in immunocompromised and in some immunocompetent hosts [4].
Early and accurate diagnosis of IFIs are important for several reasons, including timely initiation of antifungal therapy and to decrease the unnecessary use of toxic antifungal agents [5,6]. Unfortunately, the lack of specific symptoms and signs continues to be a major obstacle to the successful IFI treatment, and currently there is a lack of sensitive and specific methods for the early diagnosis of IFIs.
Currently, commonly used diagnostic methods, including fungal microscopy and culture, histopathologic detection, and biopsy, constitute the gold-standard methods of infection detection, but these methods are far from ideal [4]. Therefore,use of serological-detection methods for the early diagnosis of fungal antigens associated with IFIs continues to increase as an attractive technique to augment standard diagnostic tools [7]. Alternative serological-detection techniques include the detection of specific antigens using commercially available tests to detect galactomannan and 1, 3 β-D-glucan and genomic DNA sequencing [8]. Such indirect tests (the G test to detect the fungal cell-wall component (1-3)-β-D-glucan and the GM test to detect the Aspergillus cell-wall component galactomannan) are noninvasive, easy to handle, and can be conducted within hours. Although the value and significance of these methods are apparent, they suffer from several disadvantages due to their time-consuming nature, requirement of large quantities of both sample and reagents, and inability to produce simultaneous results.
.Low-density microarrays allow rapid, simple, and parallel detection of many elements in a single assay that can be used to perform high-throughput detection and offer great potential for disease diagnosis [9-11]. Multiple antigens can be immobilized on a slide or a membrane as capture probes to facilitate the monitoring of a number of antibodies simultaneously. Previous studies showed that Candida albicans enolase (Eno) and fructosebisphophate aldolase (Fba1) are major immunodominant antigens in IC patients and animal models, whereas Aspergillus thioredoxin reductase (TR) is the immunodominant antigen in patients infected with Aspergillus [12-14]. Here, we developed a low-density microarray assay by immobilizing these three fungal proteins onto a nitrocellulose filter and investigated the performance of serological diagnosis of invasive infection with Candida spp. and A. fumigatus.
Materials and Methods
Generation of recombinant protein
Recombinant Eno, Fba1, and TR were prepared by genetic engineering technology using methods previously described [14,15]. The recombinant plasmids were transformed into Escherichia coli BL21 (DE3) cells (Novagen; Merck Millipore, Billerica, MA, USA). Expression of recombinant antigens was induced by isopropyl-β-D- thiogalactopyranoside, and 6x Histagged recombinant proteins were purified from cell-free supernatants by chromatography on Ni2+-nitrilotriacetic acidagarose. The nitrocellulose membrane, human IgG, colloidal gold-labeled goat anti-human IgG, protein-microarray spotting instrument, and PBTX-protein microarray scanner were provided by Nanjing Potomac Biotechnology Co. Ltd. (Nanjing, China).
Serum collection
All patients were admitted to Jingling Hospital, Nanjing, China. Serum samples were collected from 105 clinically diagnosed IC patients and 42 patients with culture- and/or histology documented IA from August 2010 to January 2015. According to enzyme-linked immunosorbent assay (ELISA) results, most of the patients exhibited strong antibody reactivity to Eno, Fba1, and TR. Clinically diagnosed IC was defined as the isolation of Candida spp. from the bloodstream in at least one blood culture, and proven IA patients were classi?ed based on the standardized IFIs Group of the European Organization for the Research and Treatment of Cancer Mycoses Study Group case de?nitions [16]. Serum samples from healthy volunteers and patients without clinical or microbiological evidence of IC or IA were evaluated as controls to provide data on assay speci?city. Fifty serum samples were collected from people with Candida colonization (positive Candida spp. culture from sputum only), 35 serum samples were collected from bacteremia patients or patients with other fungal infections, and 183 serum samples were collected from healthy controls. All blood samples were centrifuged within 24 h and stored at −70°C until use. The study protocol was approved by the Ethics Committee of Jingling Hospital, and informed consent was obtained from all patients included in the study.
Protein-microarray assay
The protocols used in the microarray assays for serum testing were optimized in our laboratory and included loading concentration and loading buffer for the three antigens, antigenimmobilized conditions, blocking buffer, and dilution of patient sera. According to the optimal conditions, protein-microarray devices were constructed by Nanjing Potomac Biotechnology Co. Ltd.
For serum testing, 300 μL washing buffer (0.01 M phosphatebuffered saline with 0.1% (v/v) Tween-20 (PBS-T) (pH 7.4)) was applied to the reaction chamber to moisten the nitrocellulose membranes, followed by the application of 80 μL serum for antibody-antigen reaction. After serum passed through the nitrocellulose membrane, it was rinsed with 300 μL washing buffer, followed by the addition of 400 μL immunogold (colloidal gold-conjugated secondary antibody). After immunogold passed through the membrane, excess immunogold antibody was washed with 400 μL washing buffer, and after 20 min, the microarray devices were loaded into the PBT-X4 scanner and analyzed using the software Smartboy 3.0 (Nanjing Potomac Biotechnology Co. Ltd.). Red spots on the membrane indicating antibody-antigen reactions were scanned by the microarray scanner and converted to gray values using the software.
Statistical analysis
Variables without a normal distribution were expressed as median and interquartile ranges (25th–75th), and comparisons between two groups were performed using the Mann–Whitney U test. The potential of the three specific antibodies to be used as diagnostic markers was tested using receiver-operating characteristic (ROC) curves with calculated area under the ROC curves (AUC). The chi-squared test was used to compare the two detection methods. All tests were two-tailed, and a P<0.05 was considered statistically significant. Statistical analysis was performed using SPSS 18.0 for Windows (SPSS, Chicago, IL, USA).
Results
Optimization of reaction conditions
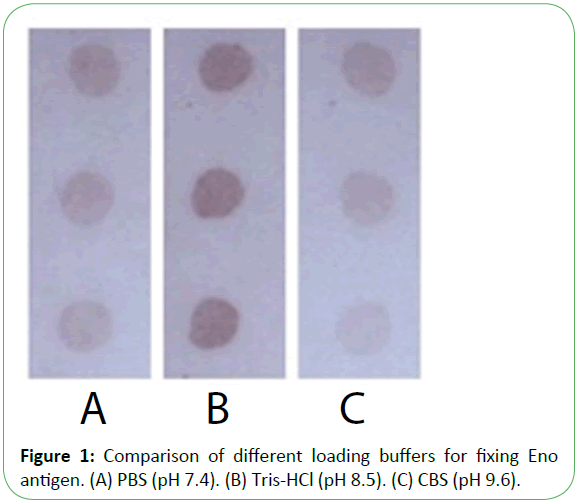
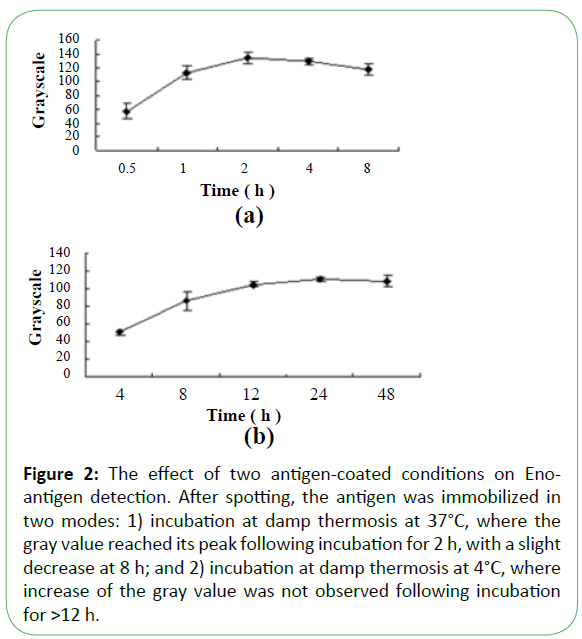
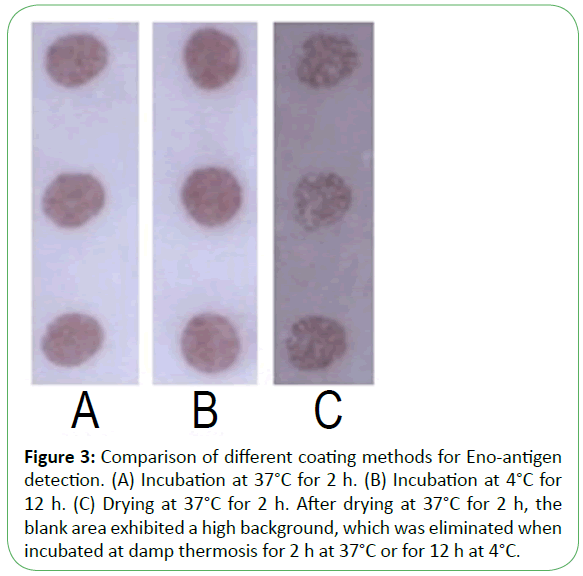
Purified recombinant Eno, Fba1, and TR were diluted with three buffers as shown in Figure 1, showing that immobilization using Tris-HCl (14.7 mL 0.1 mol/L HCl added to 50 mL 0.1 mol/L Tris, pH 8.5) was superior to results observed using PBS (NaCl 8 g, Na2HPO4·12H2O 2.08 g, KCl 0.2 g, KH2PO4 0.2 g, add distilled water to 1 L, pH7.4) or citrate-buffered saline (Na2CO3 1.59 g, NaHCO3 2.93 g, add distilled water to 1 L, pH 9.6). Following antigen spotting, the nitrocellulose membrane was incubated and antigens were blocked in three modes (Figure 2 and Figure 3). Our optimized conditions indicated that immobilization at damp thermosis for 2 h at 37°C and blocking with 5% skimmed milk powder (50 g skimmed milk powder dissolved in 1 L distilled water) for 1 h achieved suitable results. Antigen-loading concentrations and serum dilution were optimized using checkerboard titration. Results showed that the optimal loading concentrations of Eno, Fba1, and TR were 0.5 mg/mL, 0.04 mg/mL, and 1 mg/mL, respectively, with optimal serum dilutions at 1:10.
Figure 2: The effect of two antigen-coated conditions on Enoantigen detection. After spotting, the antigen was immobilized in two modes: 1) incubation at damp thermosis at 37°C, where the gray value reached its peak following incubation for 2 h, with a slight decrease at 8 h; and 2) incubation at damp thermosis at 4°C, where increase of the gray value was not observed following incubation for >12 h.
Figure 3: Comparison of different coating methods for Eno-antigen detection. (A) Incubation at 37°C for 2 h. (B) Incubation at 4°C for 12 h. (C) Drying at 37°C for 2 h. After drying at 37°C for 2 h, the blank area exhibited a high background, which was eliminated when incubated at damp thermosis for 2 h at 37°C or for 12 h at 4°C.
Specificity, stability of microarray
Specificity: Blocking tests were conducted using serum detected as strongly positive for the antigens of interest according to ELISA. Sera were diluted to 1:10 and incubated with recombinant antigens at different concentrations (0, 0.5, 1, 2, or 4 mg/mL) for 1 h at 37°C, and after drying, the microarray was scanned. The results suggested that when the final concentrations of recombinant Eno, Fba1, and TR reached 1 mg/mL, 0.5 mg/mL, and 2 mg/mL, respectively, the gray values from the scanned results were successively reduced from 188 to 33, 130 to 13, and 166 to 12, respectively indicating that the testing results exhibited high specificity. All these results indicated the microarray devices leading to good repeatability.
Stability: Protein-microarray devices using the same samples preserved at 4°C under vacuum conditions were used to test antigen-positive sera and mixed sera from controls every 15 days, and the results were compared to those from freshly prepared devices scanned within 24 h. The results indicated that the sensitivity and specificity of the test decreased gradually along with increased storage duration.
Diagnostic value of the antibody test on IC and IA patients
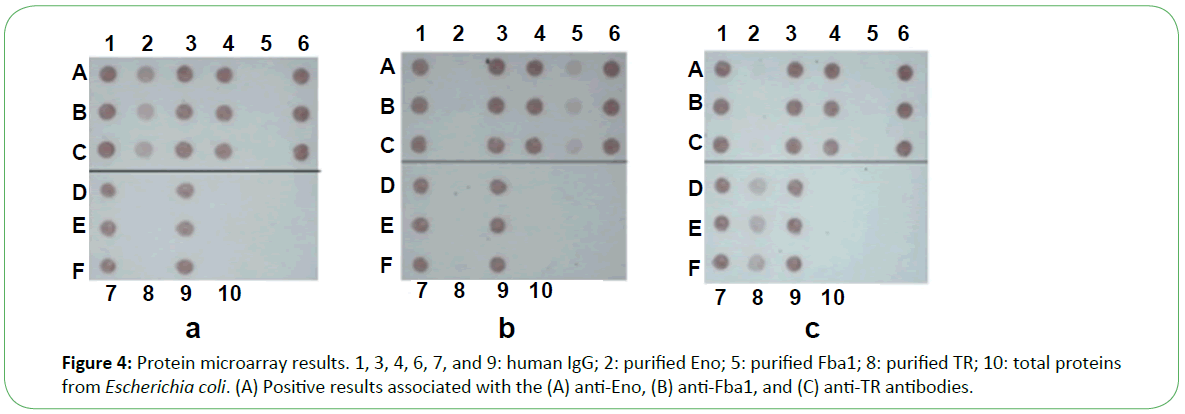
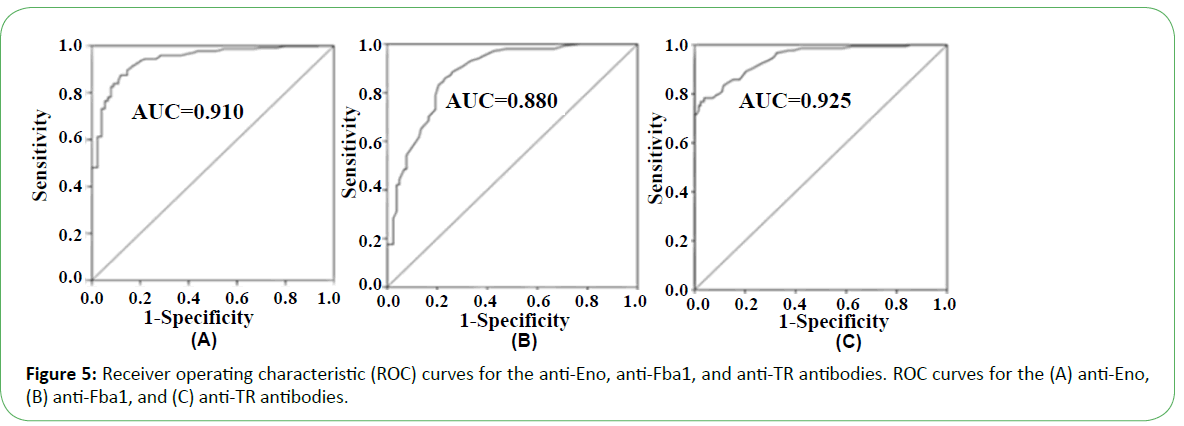
In the presence of patient sera, the antigen spot on the device was red, indicating the presence of the corresponding antibody (Figure 4). Based on the performance characteristics of the anti- Eno, -Fba1, and -TR antibodies, antigen detection in sera were assessed by ROC-curve analysis. AUC cutoff values of 66.5 and 29.5 for anti-Eno and -Fba1 antibodies in the serum samples were used to diagnose IC, returning 0.910 (95% con?dence interval (CI), ~0.899–0.920) and 0.880 (CI, ~0.870–0.890), respectively (Figure 5A and Figure 5B). At cutoff values of 66.5 and 29.5, the specificity and sensitivity of the anti-Eno and -Fba1 antibodies were 88% and 87% and 77% and 86%, respectively. At a cutoff of 58.5 for the anti-TR antibody in the serum samples used to diagnose IA, the AUC was 0.925 (95% CI, ~0.916–0.934) (Figure 5C), with a sensitivity of 78% and a speci?city of 96%. These results indicated that this method displayed superior diagnostic ability.
Testing results using the anti-Eno and anti-Fba1 antibodies in serum from IC patients
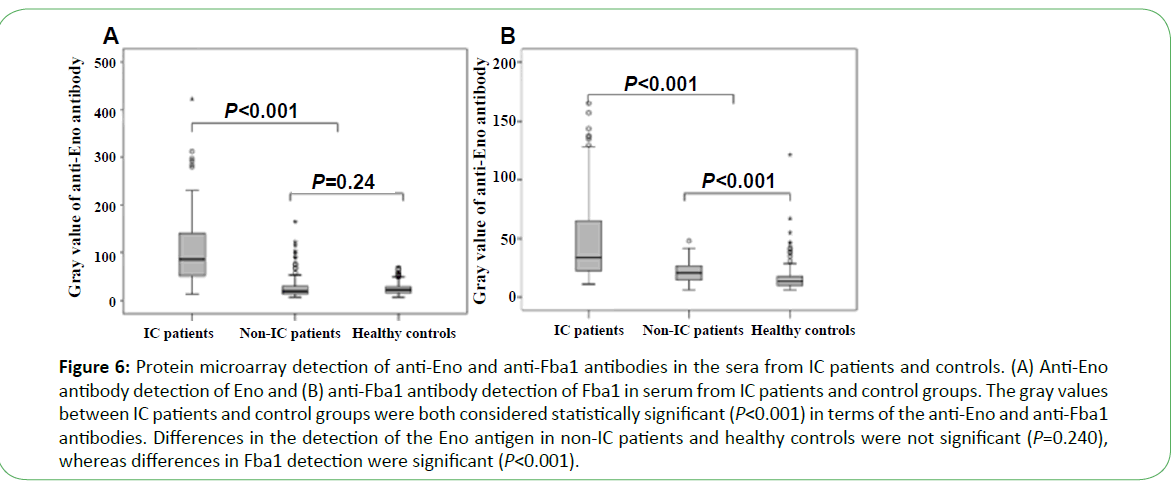
Serum from 105 patients with IC, 127 non-IC patients (including 42 IA patients, 50 cases of Candida colonization, 30 cases of bacteremia, and 5 patients with other fungal infections), and 183 healthy people was tested using the microarray devices. The anti-Eno antibody was detected in 71 cases (67.6%), with median gray values of 85 (49.5, 142) as compared with those from healthy control and non-IC patients (19 (13, 30.25), P<0.001 and 21.5 (16, 29), P<0.001, respectively). The median gray value for 68 cases (64.8%) using the anti-Fba1 antibody was 36 (23.5, 72.5) as compared with those from healthy controls and non- IC patients (13.5 (10.0, 18.0), P<0.001) and 21.0 (15.0, 27.0) P<0.001, respectively) (Figure 6). The positive rates of antibody detection of all forms of Candida infection are presented in Table 1. The positive rates for antibody detection in sera from IC patients were significantly different from that observed for Candida colonization (P<0.001). The positive rates for the anti-Eno antibody in sera from patients infected with C. albicans and non-C. albicans were 76.9% (20/26) and 64.6% (51/79), respectively ( χ2=1.37, P>0.05), and for the anti-Fba1 antibody were 65.4% (17/26) and 64.6% (51/79), respectively ( χ2=0.006, P>0.05). Additionally, compared with detection by anti-Eno or anti-Fba1 only, sensitivity and specificity improved to 81.0% and 89.0%, respectively, with positive- and negative-predictive values of 71.4% and 93.2%, respectively. When detection was undertaken using both antibodies, the results revealed higher sensitivity and negative-predictive values (Table 2).
Figure 6: Protein microarray detection of anti-Eno and anti-Fba1 antibodies in the sera from IC patients and controls. (A) Anti-Eno antibody detection of Eno and (B) anti-Fba1 antibody detection of Fba1 in serum from IC patients and control groups. The gray values between IC patients and control groups were both considered statistically significant (P<0.001) in terms of the anti-Eno and anti-Fba1 antibodies. Differences in the detection of the Eno antigen in non-IC patients and healthy controls were not significant (P=0.240), whereas differences in Fba1 detection were significant (P<0.001).
| Species | Anti-Eno antibody | Anti-Fba1 antibody | ||||||
|---|---|---|---|---|---|---|---|---|
| IC patients | Candida colonization | IC patients | Candida colonization | |||||
| n | Positive cases (%) | n | Positive cases (%) | n | Positive cases (%) | n | Positive cases (%) | |
| Candida albicans | 26 | 20(76.9) | 37 | 6(16.2) | 26 | 17(65.4) | 37 | 3(8.1) |
| Candida tropicalis | 19 | 9(47.4) | 5 | 1(20.0) | 19 | 10(52.6) | 5 | 3(60.0) |
| Candida parapsilosis | 22 | 18(81.8) | — | — | 22 | 13(59.1) | — | — |
| Candida glabrata | 10 | 7(70.0) | 3 | 2(66.7) | 10 | 10(100.0) | 3 | 1(33.3) |
| Candida guilliermondii | 5 | 2(40.0) | — | — | 5 | 3(60.0) | — | — |
| Other | 23 | 15(65.2) | 5 | 0(0) | 23 | 15(65.2) | 5 | 0(0) |
| Total | 105 | 71(67.6) a | 50 | 9(18.0) | 105 | 68(64.8) b | 50 | 7(14.0) |
Positive rates were compared against those of the respective Candida-colonization group ( χ2a =33.39, P<0.001; χ2b =34.95, P<0.001).
Table 1. Serological and microbiological surveillance of patients with IC and Candida colonization (n (%)).
| Detect anti-Eno antibody only | Detect anti-Fba1 antibody only | Combined detection | |
|---|---|---|---|
| True negative | 299 | 280 | 276 |
| False positive | 11 | 30 | 34 |
| True positive | 71 | 68 | 85 |
| False negative | 34 | 37 | 20 |
| Sensitivity (%) | 67.6 | 64.8 | 81 |
| Specificity (%) | 96.5 | 90.3 | 89 |
| NPV(%) | 89.8 | 88.3 | 93.2 |
| PPV (%) | 86.5 | 69.3 | 71.4 |
NPV: Negative-Predictive Value; PPV: Positive-Predictive Value
Table 2. Diagnostic value of anti-Eno and anti-Fba1 on the test cohort.
Testing results using the anti-TR antibody in serum from IA patients
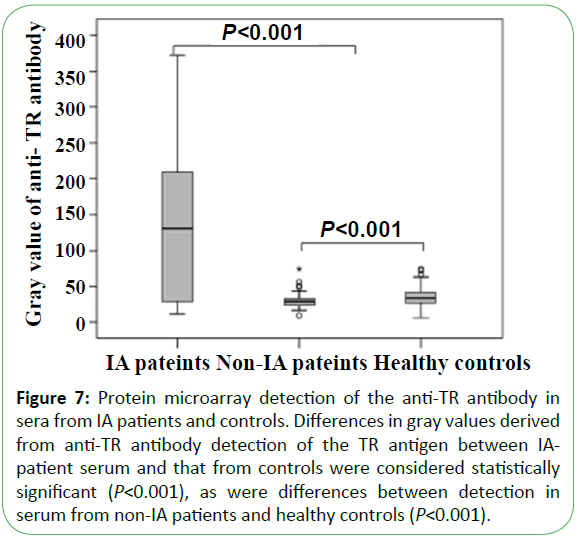
Serum from 42 IA patients, 190 non-IA patients (including 105 IC patients, 50 cases of Candida colonization, 30 cases of bacteremia, and 5 patients with other fungal infections), and 183 healthy individuals was tested using the microarray devices. The results in sensitivity and specificity of antibody detection were 71.4% (30/42) and 98.1% (366/373), respectively. Thirty cases (71.4%) were detected using the anti-TR antibody, with a median gray value of 130.0 (25.8, 218.8), whereas the median gray values for sera from healthy controls and non-IA patients at 28.0 (23.0, 32.0; P<0.001) and 33.0 (26.0, 42.0; P<0.001), respectively (Figure 7). A total of 42 patients were detected by Aspergillus galactomannan simultaneously, with 22 positive cases at a sensitivity of only 52.4%, which was significantly lower than that returned using the microarray chip to detect the anti-TR antibody (P<0.010).
Figure 7: Protein microarray detection of anti-Eno and anti-Fba1 antibodies in the sera from IC patients and controls. (A) Anti-Eno antibody detection of Eno and (B) anti-Fba1 antibody detection of Fba1 in serum from IC patients and control groups. The gray values between IC patients and control groups were both considered statistically significant (P<0.001) in terms of the anti-Eno and anti-Fba1 antibodies. Differences in the detection of the Eno antigen in non-IC patients and healthy controls were not significant (P=0.240), whereas differences in Fba1 detection were significant (P<0.001).
Discussion
The clinical diagnosis of IFI, a common infection that can cause death in immune-compromised patients, remains difficult. Recently, detection of fungal infections by serological methods through the recognition of fungal antigens, antibodies, and metabolites, has become an attractive field of study [17]. ELISA is currently used as a common serological-detection method; however, this method is time-consuming, labor-intensive, and requires large quantities of sample. Compared with the traditional methods, protein microarrays based on low-density microarray devices are advantageous because [18,19]: 1) they require small sample volumes and exhibit detection at the nanogram level; 2) they enable high-throughput, parallel analysis of thousands of protein samples within a single experiment; and 3) they exhibit a high signal-to-noise ratio.
Generally, expensive scanning equipment is essential for proteinmicroarray assays; however, the immune colloidal-gold technique avoids this limitation [20]. In this study, protein-microarray devices were constructed and optimized in combination with colloidal gold, thereby precluding the need for expensive scanning equipment.
Based on our optimized conditions, we prepared the proteinmicroarray devices to detect antigens in sera from IC and IA patients using three specific antibodies. Our results indicated that the methodology exhibited excellent specificity, repeatability, and stability and would meet the requirements of a clinical laboratory. These findings provide a foundation for further research in clinical settings to evaluate the technique for clinical applications.
Blood culture was used as the gold standard to assess the detection of sera from clinical patients using microarray devices. The positive detection rates for anti-Eno and -Fba1 antibodies were 67.6% (71/105) and 64.8% (68/105), respectively, for all Candida spp. in IC-patient sera and 71.4% (30/42) for anti-TR antibody in sera from 42 IA patients. Compared with ELISA, using microarray devices to detect anti-Eno, anti-Fba1, and anti-TR antibodies individually resulted in decreased sensitivity; however, combined detection of both anti-Eno and anti- Fba1 antibodies simultaneously increased the sensitivity to 81%. Additionally, compared to previous ELISA results, the total coincidence rates associated with the protein-microarray method for the detection of anti-Eno and anti-Fbal antibodies were 89.5% and 77.1%, respectively, with positive-coincidence rates of 89.5% and 83.6%, respectively, and negative-coincidence rates of 89.7% and 96.0%, respectively. Additionally, the consistency coefficient Kappa values were 0.751 and 0.685, respectively. The total compliance rate of these two methods for detection of the anti-TR antibody was 88.1%, with a positive-coincidence rate of 88.2%, a negativecoincidence rate of 87.5%, and a consistency coefficient Kappa of 0.662. These results indicated that the two methods were consistent in the determination of anti-Eno, anti-Fba1, and anti-TR antibodies, suggesting that the detection of dominant antibodies from Candida and Aspergillus infections by protein microarray can also provide rapid and effective etiological diagnosis of IFI.
Our results indicated that joint detection improved diagnostic sensitivity, but reduced specificity, which is required to exclude false-positive diagnosis. Suspicious positive results derived from the application of low-density protein microarrays should be retested using ELISA to confirm test results. However, all methods should be combined with investigation of patient medical history and laboratory, radiological, and histopathologic examinations to ensure a correct diagnosis.
Conclusion
This study described a new diagnostic method for detecting associated antibodies from Candida and Aspergillus infections, providing a foundation for future work in this area. Additionally, some previously reported immunodominant antigens capable of use in the diagnosis of IFI, such as histone methyltransferase fragment, dipeptidyl aminopeptidase VDPPV) and so on [21,22], were not included in this microarray device; therefore, this method requires further improvement by increasing the use of other dominant antigens to enhance diagnostic sensitivity and accuracy.
Acknowledgments
This work was supported financially by the Jiangsu Science Foundation (BE2009673).
Author contributions
XC and CFM contributed to the analysis and interpretation of data and drafted the manuscript. FQL conceived, coordinated, and designed the study, LNS and YAH performed the experiments and was involved in drafting the article. All the authors have read and approved the final manuscript.
References
- Morrissey CO (2013) Advancing the Field: Evidence for New Management Strategies in Invasive Fungal Infections. Curr Fungal Infect Rep 7:51-58.
- De Bernardis F, Arancia S, Sandini S, Graziani S, Norelli S (2015) Studies of Immune Responses in Candida vaginitis. Pathogens 4:697-707.
- Singh S,Fatima Z,Hameed S (2015) Predisposing factors endorsing Candida infections.Infez Med 23:211-213.
- Arvanitis M, Mylonakis E (2015) Diagnosis of Invasive Aspergillosis: Recent Developments and Ongoing Challenges. Eur J Clin Invest 45:646-652.
- Sipsas NV, Kontoyiannis DP (2012) Invasive fungal infections in patients with cancer in the intensive care unit. Int J Antimicrob Agents 39:464-471.
- Farmakiotis D, Kontoyiannis DP (2015) Emerging Issues With Diagnosis and Management of Fungal Infections in Solid Organ Transplant Recipients. Am J Transplant 15:1141-1147.
- Mutschlechner W, Risslegger B, Willinger B (2015) Bronchoalveolar Lavage Fluid (1,3)β-D-Glucan for the Diagnosis of Invasive Fungal Infections in Solid Organ Transplantation:A Prospective Multicenter Study. Transplantation99:e140-e144.
- Miceli MH, Maertens J (2015) Role of Non–Culture-Based Tests, with an Emphasis on Galactomannan Testing for the Diagnosis of Invasive Aspergillosis. SeminRespirCrit Care Med 36:650-661.
- Templin MF, Stoll D, SchwenkJM, Pötz O, Kramer S, et al. (2003) Protein microarrays: promising tools for proteomic research. Proteomics 3:2155-2166.
- Poetz O, SchwenkJM, Kramer S, Stoll D, Templin MF, et al. (2005) Protein microarrays: catching the proteome.Mech Ageing Dev 126:161-170.
- Richens JL, Lunt EA, O'Shea P (2015)Optimisation of proteinmicroarray techniques for analysis of the plasma proteome: Minimisation of non-speci?c binding interactions. IntImmunopharmacol 24:166-168.
- Laín A, Elguezabal N, Amutio E (2008) Use of recombinant antigens for the diagnosis of invasive candidiasis. ClinDevImmunol 2008:721-950.
- Shi LN, Li FQ, Lu JF (2012) Antibody specific to thioredoxin reductase as a new biomarker for serodiagnosis of invasive aspergillosis in non-neutropenic patients. ClinChimActa 413:938-943.
- Li FQ, Ma CF, Shi LN (2013) Diagnostic value of immunoglobulin Gantibodies against Candida enolase and fructose-bisphosphatealdolase for candidemia. BMC Infect Dis 13:253.
- Shi LN, Li FQ, Huang M (2012) Immunoproteomics based identification of thioredoxin reductase GliT and novel Aspergillusfumigatus antigens for serologic diagnosis of invasive aspergillosis. BMC Microbiology12:11.
- De Pauw B,Walsh TJ, Donnelly JP (2008) Revised de?nitions of invasive fungal disease from the European Organization for Research and Treatment of Cancer/Invasive Fungal Infections Cooperative Group and the National Institute of Allergy and Infectious Diseases Mycoses Study Group (EORTC/MSG) Consensus Group. Clin Infect Dis 46: 1813-1821.
- Sendid B, Caillot D, Baccouch-Humbert B (2003) Contribution of the Platelia Candida-specific antibody and antigen tests to early diagnosis of systemic Candida tropicalis infection in neutropenic adults. J ClinMicrobiol41:4551-4558.
- Zhu H, KlemicJF, Chang S (2000) Analysis of yeast protein kinases using protein chips. Nat Genet26:283-289.
- Cahill DJ (2001) Protein and antibody arrays and their medical applications. J Immunol Methods250:81-91.
- Zhao YH, Wang XR, Chen PC (2015) Detection of Antibodies against Avian Influenza Virus by Protein Microarray using Nucleoprotein Expressed in Insect Cells. J Vet Med Sci 77:413-419.
- Clancy CJ, Nguyen M-L, Cheng S (2008) Immunoglobulin G responses to a panel of Candida albicans antigens as accurate and early markers for the presence of systemic candidiasis. J ClinMicrobiol46:1647-1654.
- Kumar A, Ahmed R, Singh PK, ShuklaPK (2011) Identification of virulence factors and diagnostic markers using immunosecretome of Aspergillusfumigatus. J Proteomics 74:1104-1112.
Open Access Journals
- Aquaculture & Veterinary Science
- Chemistry & Chemical Sciences
- Clinical Sciences
- Engineering
- General Science
- Genetics & Molecular Biology
- Health Care & Nursing
- Immunology & Microbiology
- Materials Science
- Mathematics & Physics
- Medical Sciences
- Neurology & Psychiatry
- Oncology & Cancer Science
- Pharmaceutical Sciences