Characterization of Bioactive Compounds of Antimicrobial Metabolites Extracted from Soil Fungi
John Olayiwola1, Afolake Olanbiwoninu1, Kelechi Oleru1, Bukola Popoola1, Abiala Moses2
1Department of Biological Sciences, Ajayi Crowther University, Oyo State, Nigeria
2Department of Biological Sciences, Mountain Top University, Prayer City, Nigeria
- *Corresponding Author:
- John Olayiwola
Department of Biological Sciences, Ajayi Crowther University, Oyo State, Nigeria
E-mail: mailolusolajohn97@ yahoo.com
Received Date: November 19, 2021; Accepted Date: December 21,2021; Published Date: February 1, 2022
Citation: Olayiwola J, Olanbiwoninu A, Oleru K, Popoola B, Moses A (2021)C haracterization of Bioactive Compounds of Antimicrobial Metabolites Extracted from Soil Fungi. Med Mycol Open Access Vol:7 No:6
Abstract
Background
The global spread of bacterial resistance has significantly contributed to the increased mortality and morbidity of patients clinically due to the shortage of suitable and potent antibiotics. The concept of searching for new antibiotics in this era of antibiotic resistance pandemic remains vital therefore fungi inhabiting uncultivated soil was examined for the antimicrobial activity against multidrug-resistant bacteria.
Methods
Soil samples were collected from uncultivated farmland for fungi isolation. The isolation was done using Potato Dextrose Agar (PDA) and secondary metabolites of fungi were extracted using ethyl acetate as the solvent and the extract was dissolved with DMSO. The occurrence of bioactive compounds was carried out using GC-MS analysis. The antimicrobial susceptibility assay for the metabolites was carried out using the disc diffusion technique and zones of inhibition were measured with a vernier caliper.
Findings
The preliminary antimicrobial screening was carried out on two hundred and fifty-six (256) fungal isolates. Only forty-three (43) of the isolates were able to exhibit antimicrobial capabilities and they were identified to belong to the genera of Aspergillus and Penicillium. The pathogens used as test organisms were multidrug resistance with the maximum mean zone of inhibition to be 25.3±4.619. Crude extract of the fungi was active against all the pathogens with the maximum mean of a zone of inhibition to be 26.0±0.000 however the minimum was 14.0±0.000 zone of inhibition. The activity of the extracted metabolites ranges from 14.0±0.000 to 22.0±0.000 zone of inhibition.
Conclusion
There was the presence of different bioactive compounds like aromatic compounds, terpenes, steroids in the fungal metabolites. Fungi still constitute vital sources of antimicrobial substances and subsequently generate potential antibiotics for the treatment of diseases.
Keywords
Fungi; Metabolites; Bioactive compounds; Novel; Antimicrobials
Introduction
Soil is an ecological niche for fungi which are mainly producers of several useful bioactive natural products. Fungal secondary metabolites are rich in bioactive compounds which are highly diverse in chemistry and biological activity [1]. These fungal secondary metabolites are inexhaustible sources of new antibacterial substances. Species in the genera of the Penicillium and Aspergillus are globally distributed and are saprophytes. These genera are important sources of bioactive compounds that hold potential for novel drug substances in medicine [2].Secondary metabolites are biologically active organic compounds that are not required for normal growth, development, or reproduction; however provide a competitive advantage to the producing organism in the ecological niche [3].
Endophytic fungi due to the production of secondary metabolites have been reported as growth promoters, stress tolerance, immunity to drought, repellent of insects and herbivores [4]. The first reported antibiotics were from fungi and currently, metabolites of fungi still hold potentials for novel antimicrobials [5]. Microbial metabolites are well known to be rich sources of new potential therapeutic drugs [6]. Out of thousands of antibiotics inherent in fungi, only about 100 antibiotics have been adequately documented in the treatment of humans and animal diseases.
Bacteria have emerged as essential pathogens associated with health care for decades however, there is a continuous emergence of drug and multidrug-resistant pathogens [7], which demands the continuous search for new and highly potent antimicrobial compounds. Resistance of pathogenic microorganisms to antibiotics has become a major threat to the health sector thereby reducing the effectiveness and economic loss of the clinically available drugs. The rapid spread of Vancomycin-Resistant Enterococci (VRE) has been of particular concern around the world [8]. The resistance pressure on the bacterial population due to the constant use of antibiotics without adding novel ones has led to the emergence of resistant bacteria. Resistance to antibiotics belonging to β lactam antibiotics, quinolones, aminoglycosides, nitrofurantoin and others have also been reported [9]. Drug resistance in bacteria has become a global problem and there is an urgent and continuing search for new antibacterial agents [10] hence, the primary objective of the current study was to harness the antimicrobial potentials of the extract of fungi that has antibacterial properties along with the bioactive components.
Materials and methods
Sample Collection and Fungi Isolation
This study was carried out in Oyo town, which is in the South-Western part of Nigeria. The town is situated on latitude 8 °00 north of the equator and longitude 4 °00 east. Soil samples were randomly collected aseptically at a depth of 5 cm from 20 different locations. 1 g of the soil was suspended in 9 mL of sterile distilled water and serial dilution was carried out to reduce the load. The fungal isolates were cultured using Potato Dextrose Agar (PDA) and incubated for 5 days at 28 °C. Pure cultures of fungal isolates were identified using both macroscopic (cultural) and microscopic (morphological) features [11].
Extraction of secondary metabolites from pure isolates
The fungal isolates showing great antibacterial activities during the primary screening were selected for secondary screening using the modified method of [12]. The fungal isolates were inoculated into 250 mL Erlenmeyer flasks containing 100 mL Potato Dextrose Broth and incubated at room temperature for 21 days under stationary conditions with intermittent shaking. The broth culture was filtered to separate the mycelial and filtrate. After which equal volume of ethyl acetate was added to the filtrate, mixed well for 10 min and kept for 5 min till the two immiscible layers formed. The upper layer of ethyl acetate containing the extracted compounds was separated using a separating funnel. The extract was concentrated by removing the solvents under room temperature thereby allowing the solvent to evaporate and leaving the crude extracts. The extract was dissolved in DMSO and stored at 4 °C.
Gas chromatography and mass spectroscopy (GC-MS) analysis of bioactive compounds
Bioactive compounds in the cell-free extract of the fungi isolates that produced metabolites with higher antifungal activity against the test bacterial isolates were identified by GC-MS analysis. Briefly, 100 µL of each cell-free extract was mixed with ethyl acetate at a ratio of 1:1 and was loaded into GC-MS apparatus for analysis using the protocol of Sengupta et al. 2015 with slight modifications. The analysis was conducted using Agilent 7890
A gas chromatograph equipped and coupled to Agilent 5975 C Mass Spectrometer with a fused HP-5MS 5 % Phenyl Methyl Silox (30 m x 0.25 mm ID x 0.25 μm of the capillary column). Helium gas was used at a constant flow rate of 1.0 mL/min-1; and a fixed inlet temperature (285 °C); injection volume, 1 μL. The oven temperature program was set to an initial temperature of 90 oC, then 3 °C min-1 ramp to 180 °C and held for 10 min. The ionization voltage used will be 70 eV while a scan of 0.6 s will be applied, covering a mass range from 50 to 500 amu and the major constituents identified.
Bacterial Identification
Bacterial isolates were obtained from the Laboratory of Food Microbiology, Ajayi Crowther University and sub-cultured onto nutrient agar and incubated at 37 °C for 24 h to obtain a pure culture. The isolates were identified as Bacillus cereus and Staphylococcus aureus being Gram-positive as well as Escherichia coli and Klebsiella pneumoniae as Gram-negative bacteria.
Preliminary evaluation (agar overlay method) of fungi isolates for antimicrobial activity
Primary screening of the fungal isolates was determined by the agar overlay method. Briefly, the isolated fungi grown on PDA were incubated at 25 °C for three (3) days and overlayed by a layer of soft nutrient agar (NA) (0.75 %) inoculated with bacterial isolate with 0.5 McFarland. However, control does not have fungal isolates. All plates were incubated at 25 °C for 24 h.
Antimicrobial Susceptibility Assay
Antimicrobial susceptibility assay was performed using secondary metabolites of the fungi positive to preliminary evaluation antimicrobial of the fungal antimicrobial properties [12]. Sterile Mueller Hinton Agar was poured aseptically and 1.5 x 108 CFU/mL of bacteria liquid culture in an exponential growth phase was spread onto the surface of the plate. All the culture plates were allowed to dry for about 5 min. Wells were bored on the agar surface using a cork borer and filled with the extracted fungal metabolites respectively. After 24 h, the zone of inhibition was measured with a measuring scale and compared with the DMSO as a control.
Determination of minimum inhibitory concentration (MIC) value
The antibacterial activity of the bioactive compounds of the strain was determined in terms of minimum inhibitory concentration (MIC) against gram-positive and gram-negative bacteria by using the agar well diffusion assay. The bioactive compounds were dissolved in dimethyl sulfoxide at concentrations ranging from 0 to 1000 μg/mL and used to assay against test bacteria pathogen. Determination of MIC value was carried out using the serial dilution method after the antibacterial activity of the fungal crude extracts by the standard method described by [13] with minor modification. The final concentration of the extract in each of the test tubes numbered after dilution was dispensed on already solidified Mueller Hinton Agar. Wells were bored on the agar surface using a cork borer and filled with extract bacterial metabolites and incubated at 37 °C for 24 h, then examined for a zone of inhibition. The plate in which the zone of inhibition fails to occur was the MIC of the culture.
Results
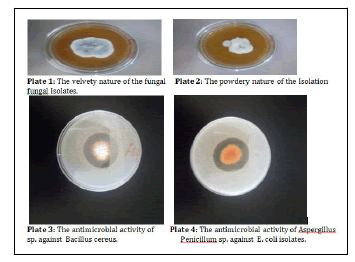
A total of two hundred and fifty-six (256) fungal isolates were screened for antimicrobial potential. Only forty-three (43) of the isolates were observed to have antimicrobial potential. They have morphological characteristics ranges from grey to brown in Penicillium spp. while cream to yellow was observed in Aspergillus spp. as shown in table 1. The fungal isolates had velvety and powdery surfaces (Plate 1 and 2).
The bacterial isolates as test organisms were identified as E. coli, Klebsiella pneumonia, Bacillus cereus and Staphylococcus aureus as shown in table 2. The antibiotics resistance profiling of the test organisms used in this study was investigated. Escherichia coli was only susceptible to tetracycline, gentamicin and chloramphenicol out of the sixteen (16) antibiotics used in the profiling. Klebsiella pneumoniae was resistance to all the antibiotics. Staphylococcus aureus was observed to be susceptible to only gentamicin however Bacillus cereus was susceptible to ampicillin and gentamicin as shown in table 3.
The main antimicrobial potential of the fungal isolates was observed through the measurement of the zone of inhibition. The metabolites of fungal isolates CUA 210, 197, 72 and 71 were able to inhibit the growth of all the pathogens used as test organisms. Metabolites of fungal isolate CUW 149 possessed activity against all the test organisms except Klebsiella pneumoniae. The antimicrobial potential of CUA 107 was observed only on the Gram-positive bacteria (Staphylococcus aureus and Bacillus cereus). Antimicrobial activity was observed to Staphylococcus aureus in assay with metabolites of fungal isolate CUA 65 while resistance was found with other test organisms (Table 4).
The minimum inhibitory concentration of the metabolites of the species of Aspergillus was observed to 421 mg/ml in the isolate CUA 197 against the pathogens (E. coli and Bacillus cereus) however it was 842 mg/mL against Klebsiella pneumonae and Staphylococcus aureus as shown in table 5. The level of antimicrobial activity of the metabolites of the isolate of Aspergillus sp. CUA 72 through two-fold dilution was 87 mg/mL for E. coli and Bacillus cereus but 174 mg/mL concentration against Klebsiella pneumonae and Staphylococcus aureus (Table 6). It was observed that mg/ml of the extract of Aspergillus sp of CUA 71 was only active against E. coli and Bacillus cereus (Table 7). The antimicrobial activity of the metabolite of Penicillium sp. CUW 149 was observed to have a minimum inhibitory concentration of 14 mg/mL against E. coli and Bacillus cereus while 28 mg/mL concentration was the MIC against Staphylococcus aureus. There was no inhibitory property against the Klebsiella pneumonae (Table 8).
GC-MS evaluation of the bioactive compounds of the fungal metabolites showed that fatty acid and ester are common bioactive compounds in the metabolites but the only aldehyde is found in the extract of isolate CUW 149 (Table 9). The bioactive compounds amine and ketone were observed in the extract of isolates CUA 71 and 72 (Table 10 and 11). The bioactive compounds obtained from isolate CUA 197 were phenol, fatty acid ester and hydrocarbons (Table 12). The GC-MS analysis showed that only metabolite of isolate CUA 210 had steroid as bioactive compound (Table 13).
| Isolates | Texture | Surface | Reverse | Zonation | Probable organism/microscopy |
|---|---|---|---|---|---|
| CUA 210 | Velvety thick | Ginger brown | Creamy brown | Concentric zones on the surface | Aspergillus sp. |
| CUW 149 | Powdery | Grey to white | Brown | Concentric zone | Penicillium sp. |
| CUA 197 | Floccose | Creamy yellow | Yellow | Furrowed | Aspergillus sp. |
| CUA 72 | Floccose | Chocolate brown | Orange | Furrowed | Aspergillus sp. |
| CUA 107 | Floccose | Creamy to brown at the centre | Orange | Furrowed | Aspergillus sp. |
| UCUW 182 | Velvety | Green to brown | Creamy dirty white | Concentric zone | Penicillium sp. |
| CUA 65 | Floccose | White to grey | Creamy | None | Penicillium sp. |
| CUA 71 | Velvety thick | White to brown | Orange to cream | Slightly furrowed | Aspergillus sp. |
Table 1: Morphological characterization of fungi.
| Locations | Escherichia coli | Klebsiella pneumonia | Bacillus cereus | Staphylococcus aureus |
|---|---|---|---|---|
| Gram reaction | - | - | + | + |
| Endospore | - | - | + | - |
| Starch hydrolysis | - | - | + | - |
| Catalase | + | + | + | + |
| Citrate | - | + | + | + |
| Motility | + | - | + | - |
| Mannitol | + | + | - | + |
| Gas | + | + | - | - |
| Oxidase | - | - | - | - |
| VP | - | + | + | + |
| Indole | + | - | - | - |
| MR | + | - | - | + |
| Glucose | + | + | + | + |
| Lactose | + | + | + | - |
Table 2: Morphological and Biochemical characterization of the Bacterial isolates.
| Antibiotic | Antibiotic disc | |||
|---|---|---|---|---|
| Escherichia coli | Klebsiella pneumonia | Staphylococcus aureus | Bacillus cereus | |
| Tetracycline | 16.0±0.000(S) | NZ | NZ | NZ |
| Erythromycin | NZ | NZ | 16.7±1.155(R) | 17.3±2.309(R) |
| Vancomycin | NZ | NZ | NZ | NZ |
| Meropenem | NZ | 17.3±2.309(R) | NZ | NZ |
| Ampicillin | NZ | NZ | NZ | 17.3±1.155(S) |
| Amoxycillin | NZ | NZ | NZ | NZ |
| Ciprofloxacin | 18.7±2.309(R) | 13.3±1.155(R) | 18.7±1.155(R) | 18.0±0.000(R) |
| Gentamicin | 16.7±2.309(S) | 14.7±1.155(R) | 14.7±1.155(S) | 18.0±0.000(S) |
| Cephalexin | NZ | NZ | NZ | NZ |
| Cefuroxime | NZ | NZ | NZ | NZ |
| Cotrimoxazole | NZ | NZ | NZ | NZ |
| Cefoperazone | NZ | 15.3±1.155(R) | NZ | NZ |
| Chloramphenicol | 20.7±1.155(S) | NZ | NZ | NZ |
| Ceftriaxone | NZ | 15.3±1.155(R) | -NZ | NZ |
| Cefotaxime | NZ | 16.0±3.464(R) | NZ | NZ |
| Amikacin | 25.3±4.619(S) | 16.0±0.000(R) | NZ | NZ |
| S= sensitive, I = intermediate and R = resistant. NZ=no zone, -=not applicable | ||||
Table 3: Antibiotic resistance profiling of test bacterial pathogens.
| Isolates | Escherichia coli | Klebsiella pneumonia | Bacillus cereus | Staphylococcus aureus |
|---|---|---|---|---|
| Inhibition zone (mm) (mean ± SD) | ||||
| CUA 210 | 19.3±2.309 | 14.0±0.000 | 15.3±2.309 | 22.0±0.000 |
| CUW 149 | 19.3±2.309 | NZ | 19.3±2.309 | 14.0±0.000 |
| CUA 197 | 20.7±4.619 | 14.0±0.000 | 16.7±2.309 | 22.0±0.000 |
| CUA 72 | 23.3±4.619 | 16.7±2.309 | 14.0±0.000 | 20.7±2.309 |
| CUA 107 | NZ | NZ | 18.0±0.000 | 22.0±0.000 |
| CUW 182 | NZ | NZ | NZ | NZ |
| CUA 65 | NZ | NZ | NZ | 18.0±0.000 |
| CUA 71 | 24.7±4.619 | 14.0±0.000 | 20.7±2.309 | 26.0±0.000 |
| NZ:no zone | ||||
Table 4: Antibacterial activities of fungal crude extract using agar well diffusion method.
| Pathogens | Minimum Inhibitory Concentration | |||||
|---|---|---|---|---|---|---|
| E. coli | 842 mg/mlmean ±S.D | 421 mg/mlmean ±S.D | 210.5 mg/mlmean ±S.D | 105.25 mg/mlmean ±S.D | 52.625 mg/mlmean ±S.D | MIC value(mg/ml) |
| 24.0 ±2.828 | 18.0 ±5.657 | - | - | - | 421 | |
| Klebsiella pneumonae | 18.0 ±0.000 | - | - | - | - | 842 |
| Staphylococcus aureus | 20.0 ±2.828 | - | - | - | - | 842 |
| Bacillus cereus | 22.0 ±0.000 | 16.0 ±2.828 | - | - | - | 421 |
Table 5: Minimum inhibitory concentration of the ethyl extract of Aspergillus sp. CUA 197.
| Pathogens | Minimum Inhibitory Concentration | |||||
|---|---|---|---|---|---|---|
| E. coli | 174 mg/mlmean ±S.D | 87 mg/mlmean ±S.D | 43.5 mg/mlmean ±S.D | 21.75 mg/mlmean ±S.D | 10.87 mg/mlmean ±S.D | MIC value(mg/ml) |
| 20.0 ±2.828 | 14.0 ±0.000 | - | - | - | 87 | |
| Klebsiella pneumonae | 18.0 ±0.000 | - | - | - | - | 174 |
| Staphylococcus aureus | 18.0 ±5.657 | - | - | - | - | 174 |
| Bacillus cereus | 22.0 ±0.000 | 16.0 ±2.828 | - | - | - | 87 |
Table 6: Minimum inhibitory concentration of the ethyl extract of Aspergillus sp. CUA 72.
| Pathogens | Minimum Inhibitory Concentration | |||||
|---|---|---|---|---|---|---|
| E. coli | 38 mg/mlmean ±S.D | 19 mg/mlmean ±S.D | 9.5 mg/mlmean ±S.D | 4.75 mg/mlmean ±S.D | 2.38 mg/mlmean ±S.D | MIC value(mg/ml) |
| 22.0 ±0.000 | 16.0 ±2.828 | - | - | - | 19 | |
| Klebsiella pneumonae | 18.0 ±0.000 | - | - | - | - | 38 |
| Staphylococcus aureus | 20.0 ±2.828 | - | - | - | - | 38 |
| Bacillus cereus | 22.0 ±0.000 | 16.0 ±2.828 | - | - | - | 19 |
Table 7: Minimum inhibitory concentration of the ethyl extract of Aspergillus sp. CUA 71.
| Pathogens | Minimum Inhibitory Concentration | |||||
|---|---|---|---|---|---|---|
| E. coli | 28 mg/mlmean ±S.D | 14 mg/mlmean ±S.D | 7 mg/mlmean ±S.D | 3.5 mg/mlmean ±S. D | 1.75 mg/mlmean ±S.D | MIC value(mg/ml) |
| 22.0 ±0.000 | 18.0 ±0.000 | - | - | - | 14 | |
| Klebsiella pneumonae | - | - | - | - | - | - |
| Staphylococcus aureus | 18.0 ±0.000 | - | - | - | - | 28 |
| Bacillus cereus | 22.0 ±5.657 | 16.0 ±2.828 | - | - | - | 14 |
Table 8: Minimum inhibitory concentration of the ethyl extract of Penicillium sp. CUW 149.
| Compound name | Molecular formula | Molecular weight | Iupac name | Nature of compound |
|---|---|---|---|---|
| 1,2-Benzenedicarboxylic acid,bis(2-methylpropyl) ester | C18H26O4 | 306.402 g/mol | Bis(2-methylpropyl)4,5-dimethylbenzene-1,2-dicarboxylate | Fatty acid ester |
| 3-Hydroxy-2-pyridin-3-yl-propenal | C8H7NO2 | 149.149 g/mol | (Z)-3-hydroxy-2-pyridin-3-ylprop-2-enal | Aldehyde |
| Benzenebutanamine | C10H15N | 149.237 g/mol | 4-phenylbutan-1-amine | Amines |
| 2H-1,4-Benzoxazin-3(4H)-one | C8H7NO2 | 149.149 g/mol | 4H-1,4-benzoxazin-3-one | Ketones |
| Dibutyl phthalate | C16H22O4 | 278.348 g/mol | Dibutyl benzene-1,2-dicarboxylate | Fatty acid ester |
| 1,2-Benzenedicarboxylic acid, butyloctyl ester | C20H30O4 | 334.456 g/mol | 1-O-butyl2-O-Octylbenzene-1,2-dicarboxylate | Fatty acid ester |
| 1,2-Benzenedicarboxylic acid, butyl2-ethylhexyl ester | C20H30O4 | 334.456 g/mol | 1-O-butyl2-O-(2-ethylhexyl)benzene-1,2-dicarboxylate | Fatty acid ester |
Table 9: Bioactive compound in the metabolite of the fungal isolate cuw 149 extracts.
| Compound name | Molecular formula | Molecular weight | Iupac name | Nature of compound |
|---|---|---|---|---|
1,2-Benzenedicarboxylic acid,bis(2-methylpropyl) ester |
C18H26O4 |
306.402 g/mol |
Bis(2-methylpropyl)4,5-dimethylbenzene-1,2-dicarboxylate |
Fatty acid ester |
3-Hydroxy-2-pyridin-3-yl-propenal |
C8H7NO2 |
149.149 g/mol |
(Z)-3-hydroxy-2-pyridin-3-ylprop-2-enal |
Aldehyde |
Benzenebutanamine |
C10H15N |
149.237 g/mol |
4-phenylbutan-1-amine |
Amines |
2H-1,4-Benzoxazin-3(4H)-one |
C8H7NO2 |
149.149 g/mol |
4H-1,4-benzoxazin-3-one |
Ketones |
Dibutyl phthalate |
C16H22O4 |
278.348 g/mol |
Dibutyl benzene-1,2-dicarboxylate |
Fatty acid ester |
1,2-Benzenedicarboxylic acid, butyloctyl ester |
C20H30O4 |
334.456 g/mol |
1-O-butyl2-O-Octylbenzene-1,2-dicarboxylate |
Fatty acid ester |
1,2-Benzenedicarboxylic acid, butyl2-ethylhexyl ester |
C20H30O4 |
334.456 g/mol |
1-O-butyl2-O-(2-ethylhexyl)benzene-1,2-dicarboxylate |
Fatty acid ester |
Table 10: Bioactive compound in the metabolite of the fungal isolate cua 71 extracts.
| Compound name |
Molecular formula |
Molecular weight |
Iupac name |
Nature of compound |
|---|---|---|---|---|
2,4-Di-tert-butylphenol |
C14H220 |
206.329 g/mol |
2,4-ditert-butylphenol |
Phenols |
Cyclododecane |
C12H24 |
168.324 g/mol |
Cyclododecane |
Alkanes |
Fluorenone oxime |
C13H9NO |
195.221 g/mol |
N-fluoren-9-ylidenehydroxylamine |
Amines |
Trifluoroacetic acid, n-tridecyl ester |
C15H27F3O2 |
296.374 g/mol |
Tridecyl 2,2,2-trifluoroacetate |
Fatty acid ester |
1,2-Benzenedicarboxylic acid, butyl 2-methylpropyl ester |
C16H22O4 |
278.348 |
1-O-butyl 2-O-(2-methylpropyl) benzene-1,2-dicarboxylate |
Fatty acid ester |
Phthalic acid, butyl oct-3-yl ester |
C20H30O4 |
334.456 g/mol |
1-O-butyl 2-O-octan-3-yl benzene-1,2-dicarboxylate |
Fatty acid ester |
1,2-Benzenedicarboxylic acid, butyl 2-methylpropyl ester |
C16H22O4 |
278.348 |
1-O-butyl 2-O-(2-methylpropyl) benzene-1,2-dicarboxylate |
Fatty acid ester |
| L-Proline, N-valeryl-, butyl ester | C14H25NO3 | 255.358g/mol | Butyl 1-pentanoylpyrrolidine-2-carboxylate | Fatty acid ester |
| 1-Penten-3-one, 4-methyl-1-(2,6,6-trimethyl-2-cycl-ohexen-1-yl)- | C15H24O | 220.356g/mol | (E)-4-methyl-1-(2,6,6-trimethylcyclohex-2-en-1-yl) pent-1-en-3-one | Terpenes |
| 2H-1-Benzopyran-2-one, 6,7-dimethoxy-4-methyl | C12H12O4 | 220.224g/mol | 6,7-dimethoxy-4-methylchromen-2-one | Aromatic |
| Dibutyl phthalate | C16H22O4 | 278.348g/mol | Dibutyl benzene-1,2-dicarboxylate | Fatty acid ester |
| Benzene, 1,2-dimethoxy-4-(1-propenyl)- | C11H14O2 | 178.231g/mol | 1,2-dimethoxy-4-[(E)-prop-1-enyl] benzene | Aromatic |
| 2-Butenoic acid, 2-cyano-3-methyl-, ethyl ester | C8H11NO2 | 153.181g/mol | Ethyl 2-cyano-3-methylbut-2-enoate | Fatty acid ester |
Table 11: Bioactive Compound in the Metabolite of the Fungal Isolate CUA 72 extracts.
| Compound name | Molecular formula | Molecular weight | Iupac name | Nature of compound |
|---|---|---|---|---|
| Cyclopentane, (2-methyl-1-propenyl)- | C9H16 | 124.227 g/mol | 2-methylprop-1-enylcyclopentane | Alkanes |
| 1,1-dimethyl-4-methylene cyclohexane | C9H16 | 124.227 g/mol | 1,1-dimethyl-4-methylidene cyclohexane | Alkanes |
| Phenol,2,6-bis(1,1-dimethy-lethyl)- | C14H22O | 206.329 | 2,6-ditert-butylphenol | Phenols |
| Cycloheptane, methyl- | C8H16 | 112.216 | Methylcycloheptane | Alkanes |
| Phthalic acid, isobutyl non-5-yn-3-yl ester | C21H28O4 | 344.451 | 1-O-(2-methylpropyl)2-O-non-5-yn-3-yl benzene-1,2-dicarboxylate | Fatty acid ester |
| Dibutyl phthalate | C16H22O4 | 278.348 g/mol | Dibutyl benzene-1,2-dicarboxylate | Fatty acid ester |
| Dibutyl phthalate | C16H22O4 | 278.348 g/mol | Dibutyl benzene-1,2-dicarboxylate | Fatty acid ester |
Table 12: Bioactive Compound in the Metabolite of the Fungal Isolate CUA 197 extracts.
| Compound name | Molecular formula | Molecular weight | Iupac name | Nature of compound |
| 2,4-di-tert-butylphenol | C14H22O | 206.329 g/mol | 2,4-ditert-butylphenol | Phenols |
| 1-Octadecene | C18H36 | 252.486 g/mol | Octadec-1-ene | Alkene |
| Phthalic acid, isobutyl nonyl ester | C21H32O4 | 348.483 g/mol | 2-O-(2-methylpropyl) 1-O-nonylbenzene-1,2-dicarboxylate | Fatty acid ester |
| Dibutyl phthalate | C16H22O4 | 278.348 g/mol | Dibutyl benzene-1,2-dicarboxylate | Fatty acid ester |
| Dibutyl phthalate | C16H22O4 | 278.348 g/mol | Dibutyl benzene-1,2-dicarboxylate | Fatty acid ester |
| Cholesterol | C27H46O | 386.664 g/mol | (3S,8S,9S,10R,13R,14S,17R)-10,13-dimethyl-17-[(2R)-6-methylheptan-2-yl]-2,3,4,7,8,9,11,14,15,16,17-dodeca-hydro-1H-cyclopenta[a]phenanthren-3-ol | Steroid |
| Cholesterol | C27H46O | 386.664 g/mol | (3S,8S,9S,10R,13R,14S,17R)-10,13-dimethyl-17-[(2R)-6-methylheptan-2-yl]-2,3,4,7,8,9,11,14,15,16,17-dodeca-hydro-1H-cyclopenta[a]phenanthren-3-ol | Steroid |
Table 13: Bioactive Compound in the Metabolite of the Fungal Isolate CUA 210 extracts.
Discussion
The search for the new potential antimicrobial substance is vital in this era of multi-drug resistant pathogenic bacteria in clinical practice. This study investigates the antimicrobial substances in the metabolites of species of Aspergillus and Penicillium isolated from uncultivated soil. The isolation of these fungi from uncultivated soil has earlier been reported by [14] which confirmed the report of our study.
The pathogens used for the antimicrobial study were identified to be E. coli and Klebsiella pneumonia as Gram-negative bacteria. The bacteria used as Gram-positive were Staphylococcus aureus and Bacillus cereus. The Klebsiella pneumoniae was resistance to all the antibiotics used for the susceptibility assay however, E. coli was only susceptible to tetracycline, gentamicin and chloramphenicol. These two Gram-negative bacteria are resistance to more than three antibiotics which showed that they are multiple antibiotic-resistant bacteria. MDR bacteria have earlier been reported among the family Enterobacteriaceae especially Klebsiella pneumoniae and E. coli which is similar to Peirano and Pitout, (2019) that there is an extensive antibiotic resistance pattern in Enterobacteriaceae. In this work, multi-drug resistance was also observed in the Gram-positive bacterial pathogens which are Bacillus cereus and Staphylococcus aureus. There have been a lot of reports on the multi-drug resistance in Staphylococcus aureus [15,16] which is similar to the findings of this work where Bacillus cereus and Staphylococcus aureus were found multi-drug resistance.
The metabolites of the species of Aspergillus and Penicillium were found to have antimicrobial properties against both Gram-negative and Gram-positive bacteria. Four species of Aspergillus were very active in respect to their antimicrobial properties in this work which correspond to [17] who reported inhibitory activity of A. niger against P. aeruginosa, S. aureus, and Bacillus sp. They were active against all the pathogens investigated except the metabolite of the Aspergillus isolate CUW 149 that was not active against Klebsiella pneumoniae. This finding is similar to the report of [18], that metabolites of Aspergillus sp. are majorly active against Staphylococcus sp. Interestingly; the species of Penicillium isolate CUA 65 was only active against Staphylococcus aureus. However, the metabolite of the species of Penicillium isolates CUW 182 did not show any activity against any of the pathogens.
The minimum concentration of the metabolites of the fungal isolates was investigated in this work and it was found that as low as 14 mg/ml concentration was active against even E. coli and Bacillus aureus in the metabolite extracted from Penicillium spp. It was noted in this work that the metabolites whose concentrations are higher were found effective against Klebsiella pneumonae and Staphylococcus aureus across the findings [21,22]. This finding confirmed the report of a previous study that reported Klebsiella pneumonae and Staphylococcus aureus as notorious is the antibiotic resistance.
The bioactive compounds of the metabolites were mainly fatty acid and ester in all the fungi investigated in this work. Along with these components, amine and ketone were found in some of the fungal metabolites however aldehyde was only in the metabolite of Penicillium isolate CUW 149. The presence of ketones and amine among the bioactive compounds in this work correspond to [19] that reported steroids and alkaloids among other bioactive compounds. The metabolite of the Aspergillus isolates CUA 210 contains a steroid which is an indication of the microbial hormone. Other bioactive compounds found in our work are terpenes, aromatic compounds, phenols, amines, etc which are similar to the report of critical review of bioactive compounds by [20].
Conclusion
The antimicrobial activity of the fungal metabolites as found in our work provide evidence of the possibility of discovering novel antimicrobials from fungi. Fungi inhabiting virgin or long time uncultivated soil could contain a lot of fungi that would be in natural form with little or no environmental alteration in their biological form.
References
- Abouelhassan Y, Garrison A T, Yang H, Chavez-Riveros A, Burch G M, et al. (2019) Recent progress in natural-product-inspired programs aimed to address antibiotic resistance and tolerance. J Med Chem 62:7618–7642
- Nielsen J C, Grijseels S, Prigent S, Dainat J, Nielsen K F, et al. (2017) Global analysis of biosynthetic gene clusters reveals vast potential of secondary metabolite production in Penicillium species. Nat Microbiol 2:17044
- Craney A, Ahmed S, Nodwell J (2013) Towards a new science of secondary metabolism. J Antibiot 66:387–400
- Thatoi H, Behera B C and Mishra R R (2013) Ecological role and biotechnological potential of mangrove fungi: A review Mycology4:4
- Willems, Thomas, Maarten L, De Mol, Wim K, et al. (2020) "Alkaloids from Marine Fungi: Promising Antimicrobials" Antibiotics 9 6:340
- Cragg G M, & Newman D J (2013) Natural products: a continuing source of novel drug leads. Biochi mic aetbio physic aacta, 1830:3670–3695
- PeiranoG,andPitoutJDD(2019)ExtendedspectrumbetalactamaseproducingEnterobacteriaceae: update on molecular epidemiology and treatment options. Drugs 79: 1529–1541
- MeleseA,GenetC&AndualemT(2020)PrevalenceofVancomycinresistantenterococci(VRE) in Etiopia: a systematic review and meta-analysis.BMC Infect Dis20:124
- Abiala M, Olayiwola J, Babatunde O, Aiyelaagbe O and Akinyemi S (2016) Evaluation of therapeutic potentials of plant extracts against poultry bacteria threatening public health. BMC Complement Altern Med 16:417
- Kibret M & Abera B (2011) Antimicrobial susceptibility patterns of E. coli from clinical sources in northeast Ethiopia. African health sciences, 11 Suppl 1(Suppl 1), 40–45
- Yingwu Shi, Kai Lou and Chun Li (2009) Isolation, quantity distribution and characterization of endophytic microorganisms within sugar beet African Journal of Biotechnology 8:835-840
- Sheeba H, Syed Ali M, Anuradha V (2019) Bioactive compounds and antimicrobial activity of fungal crude extract from medicinal plants. J Pharm Sci & Res 11:1826-1833
- Bhardwaj A, Sharma D, Jadon N and Agrawal PK (2015) Antimicrobial and Phytochemical Screening of Endophytic Fungi Isolated from Spikes of Pinus roxburghii. Arch Clin Microbiol 3:1
- Oleru K, Olanbiwoninu A, Olayiwola J and Popoola B (2021) Potential Antimicrobial Substances from the Characterized Bioactive Compounds Extracted from Secondary Metabolites of Aspergillus terreus. Res J Microbiol 16:8-18
- Gajdacs M (2019) The continuing threat of methicillin-resistant Staphylococcus aureus. Antibiotics 8:52
- Fasiku SA, Olayiwola JO, Babayemi EO, Lasekan TO and Olanbiwoninu AA (2020) Multiple Antibiotic Resistant Surveillance of Oxacillin-resistant Staphylococcus aureus Isolated from Beef and Frozen Poultry Meat (Chicken). J Appl Life Sci Int 23:29-35
- Al-Shaibani ABA, Al-Shakarchi FI, Ameen RS (2013) Extraction and characterization of antibacterial compound from Aspergillus niger. J Al-Nahrain Univ/Sci 16:167–174
- Al-Fakih A A, & Almaqtri W (2019) Overview on antibacterial metabolites from terrestrial Aspergillus spp. Mycology 10:191–209
- Jin J, Zhao Q, Zhang XM, Li WJ (2018) Research progress on bioactive products from endophytes. J Microbiol 38:103–113
- Zheng, Ruihong, Shoujie Li, Xuan Zhang, and Changqi Zhao (2021)"Biological Activities of Some New Secondary Metabolites Isolated from Endophytic Fungi: A Review Study" Int J Mol Sci 2:959
- Jose PA and Jha B (2016) New Dimensions of Research on Actinomycetes: Quest for Next Generation Antibiotics. Front Microbiol 7:1295
- Yao G, Chen X, Zheng H, Liao D, Yu Z, et al. (2021) Genomic and Chemical Investigation of Bioactive Secondary Metabolites From a Marine-Derived Fungus Penicillium steckii P2648. Front Microbiol 12:600991
Open Access Journals
- Aquaculture & Veterinary Science
- Chemistry & Chemical Sciences
- Clinical Sciences
- Engineering
- General Science
- Genetics & Molecular Biology
- Health Care & Nursing
- Immunology & Microbiology
- Materials Science
- Mathematics & Physics
- Medical Sciences
- Neurology & Psychiatry
- Oncology & Cancer Science
- Pharmaceutical Sciences