Combination of Phenotypic Tests as a Screening Approach for the Differentiation of Cryptic Species Candida albicans and Candida dubliniensis
Marcos Fabio Gadelha Rocha, Silviane Praciano Bandeira, Lucas Pereira de Alencar, Glaucia Morgana de Melo Guedes, Debora de Souza Collares Maia Castelo-Branco, Tereza de Jesus Pinheiro Gomes Bandeira, Rossana de Aguiar Cordeiro, Jose Julio Costa Sidrim and Raimunda Samia Nogueira Brilhante
DOI10.21767/2471-8521.100024
Marcos Fabio Gadelha Rocha1,2, Silviane Praciano Bandeira1, Lucas Pereira de Alencar1, Glaucia Morgana de Melo Guedes1, Debora de Souza Collares Maia Castelo-Branco1, Tereza de Jesus Pinheiro Gomes Bandeira3, Rossana de Aguiar Cordeiro1, Jose Julio Costa Sidrim1 and Raimunda Samia Nogueira Brilhante1*
1Department of Pathology and Legal Medicine, Postgraduate Program in Medical Microbiology, Specialized Medical Mycology Center, Federal University of Ceara, Fortaleza, Ceara, Brazil
2School of Veterinary Medicine, Postgraduate Program in Veterinary Sciences, State University of Ceara, Fortaleza, Ceara, Brazil
3School of Medicine, Christus College – Unichristus, Fortaleza, Ceara, Brazil
- *Corresponding Author:
- Brilhante RSN
Department of Pathology and Legal Medicine, Postgraduate Program in Medical Microbiology
Specialized Medical Mycology Center, Federal University of Ceara, Fortaleza, Ceara, Brazil
Fax: +55 85 3366-8319
E-mail: brilhante@ufc.br
Received date: October 31, 2016; Accepted date: March 08, 2017; Published date: March 18, 2017
Citation: Rocha MFG, Bandeira SP, de Alencar LP, de Melo Guedes GM, Maia Castelo-Branco DSC, et al. (2017) Combination of Phenotypic Tests as a Screening Approach for the Differentiation of Cryptic Species Candida albicans and Candida dubliniensis Med Mycol Open Access 3:24. doi: 10.21767/2471-8521.100024.
Copyright: © 2017 Rocha MFG, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Abstract
Background: Candida albicans, an important pathogen for humans and animals, shares phenotypic features with Candida dubliniensis, leading to misidentification. Thus, the goal of this study was to apply a combination of phenotypic tests for the differentiation of these cryptic species.
Methods and findings: Thirty-seven azole-resistant C. albicans from animals and 03 C. dubliniensis from humans were included in this study. Purity of strains was evaluated on CHROMagar Candida™, on which both species present green colonies. Then, phenotypic characterization was performed based on the growth pattern on sunflower seed agar, where C. albicans presents smooth colonies and C. dubliniensis presents rough colonies, and esterase production on Tween 80™ agar, which is positive for C. albicans, forming an opacification zone, and negative for C. dubliniensis. Molecular differentiation was performed with primers CALF/CALR for the amplification of ITS1/ITS2 regions of rDNA, yielding an amplicon of 100 bp for C. albicans, but not for C. dubliniensis. Of the 37 C. albicans, 35 showed green colonies on CHROMagar Candida Medium™, 36 presented smooth colonies on sunflower seed agar, while 34 showed an opacification zone on Tween 80™ agar. PCR yielded 100-bp-amplicons for all 37 C. albicans strains, confirming their identification. Control strains of C. dubliniensis showed the expected phenotypic features and amplicons were not obtained for the specific PCR reaction.
Conclusions: The phenotypic methods used were not absolutely effective, however, the combined observation of smooth colonies on sunflower seed agar with an opacification zone on Tween 80™ agar leads to a reliable presumptive phenotypic identification of C. albicans.
Keywords
Candida albicans; Candida dubliniensis; Animal; Phenotypic differentiation
Introduction
Yeasts of the genus Candida are the main fungal pathogens for humans and animals, and are important causes of nosocomial infection. Even though C. albicans is still commonly associated with superficial and deep infections, Candida infections with non-C. albicans species has increased in recent decades [1]. C. dubliniensis, for instance, was first described by Sullivan et al. [2], from oral lesions of HIV-positive patients and, since then, has been isolated from several other cases of human infections [3,4], including fungemia [5]. Isolation of this species from animals is rare, but C. dubliniensis has been recovered from bird droppings [6] and the surface of ticks that had contact with bird excrements [7].
C. albicans and C. dubliniensis are cryptic species and share several phenotypic characteristics, as germ tube formation and chlamydospore production, when grown on cornmeal agar [8]. Although some phenotypic characteristics slightly differ between these species, definite differentiation of C. albicans and C. dubliniensis relies on molecular methods [9,10].
Studies have shown that C. dubliniensis develops antifungal resistance more promptly than C. albicans [11]. However, our group has recovered azole-resistant C. albicans from several animal species [12-15]. Therefore, this study aimed to reidentify these isolates, focusing on evaluating the accuracy of combined phenotypic tests for the differentiation of C. albicans and C. dubliniensis, using the polymerase chain reaction (PCR)-based identification as gold standard.
Methods
Strains
The inclusion criteria used for the strains in this study were: previous phenotypic identification as C. albicans and resistance to azole derivatives. Thus, 37 C. albicans strains previously stored in saline at 4°C in the culture collection of the Specialized Medical Mycology Center (CEMM) of the Federal University of Ceara; Brazil, were subcultured in Sabouraud broth, and incubated at 35°C for 24 h. Then, the strains were subcultured on Sabouraud agar and incubated at 35°C, for further 24 h. After recovery, the purity of the strains was confirmed on chromogenic agar and additional phenotypic tests were performed to assure their identification as C. albicans. Three strains of C. dubliniensis from human infections and C. albicans ATCC 10231 were also included in the study as control strains.
Phenotypic methods
Germ tube test
After confirming the viability of the strains, the isolates were tested for germ tube formation. One fungal colony was inoculated in 0.5 ml of human serum and incubated at 37°C for a period of 3 h. Every 30 minutes, a drop of the suspension was observed under light microscopy at magnifications of 100x and 400x. Tubular fungal structures with parallel walls growing out of blastoconidia, without constrictions, were compatible with germ tubes. Both C. albicans and C. dubliniensis are capable of forming germ tubes and true hyphae [2].
Micromorphological analysis on cornmeal agar
The strains were seeded on cornmeal agar supplemented with Tween 80™ and incubated for 3 to 5 days, at room temperature (28ºC). Then, the plates were observed under light microscopy at magnifications of 100x and 400x, in order to analyze the microscopic arrangement of each isolate and the production of chlamydospores, which are characteristic of both C. albicans and C. dubliniensis [2].
Growth on CHROMagar Candida medium™
The strains were grown on agar CHROMagar Candida™ (Difco ™) by streaking into quadrants. After incubation at 35°C, for 48 h, the chromogenic patterns of each tested strain were observed. According to the manufacturer, colonies of C. albicans and C. dubliniensis grown on this medium are green, after incubation for 48 h [16].
Growth on sunflower seed agar
Growth on sunflower seed agar was evaluated for the phenotypic differentiation of C. albicans and C. dubliniensis. The strains were streaked, as described by Khan et al. [17], incubated at 28°C, for three days, and daily observed to evaluate the texture of the colonies. On this agar, C. albicans colonies have a smooth texture, while those of C. dubliniensis have a rough texture [17].
Esterase production on Tween 80™ agar
Strains were inoculated in duplicate on agar containing Tween 80™, as described by Slifkin [18], in order to evaluate the production of esterases. The plates were incubated at 30°C for up to 12 days and were daily observed for the presence of an opacification zone around the point of inoculation. C. albicans produces esterases, forming an opacification zone, after up to 5 days of incubation, whereas C. dubliniensis does not produce these enzymes, hence opacification zones are not observed, after 12 days of observation.
Molecular identification by polymerase chain reaction (PCR)
Strains were grown on Sabouraud agar, for 48 hours, and submitted to DNA extraction using the High Pure PCR Template Preparation Kit (Roche™). After extraction, DNA was quantified through Qubit™. The obtained DNA samples were subjected to PCR, according to the methodology described by Ahmad et al. [8], using species-specific primers to amplify specific ITS1 and ITS2 regions of rDNA. For the identification of C. albicans, CALF (5'-3'TGGTAAGGCGGGATCGCTT) and CALR (5'GGTCAAAGTTTGAAGATATAC) primers were used. The PCR reaction was performed in a final volume of 25 μL, containing 2 μL of each primer (50 pmol.μL-1), 5 μL of 1 mM dNTP mix (Promega™), 0.25 U GoTaq® Hot Start Polymerase (Promega™), 5 μL 5X Green GoTaq® Flexi Buffer (Promega ™), 5 μL MgCl2 (Promega™), 3.95 μL of ultrapure water and 2 μL of DNA template. Cycling conditions included initial denaturation at 95°C for 5 min, followed by 30 cycles at 95°C for 1 min, 55°C for 30 s and 72°C for 1 min, with final extension at 72°C, for 10 min. After the reaction, amplicons were subjected to electrophoretic run on 2% agarose gel, and 100bp-DNA bands were compatible with C. albicans.
Results
Confirmatory phenotypic tests showed that all C. albicans strains produced germ tubes in human serum and chlamydospores on cornmeal agar. On CHROMagar Candida™, 35 (94.6%) strains were green, while two (5.4%) were blue.
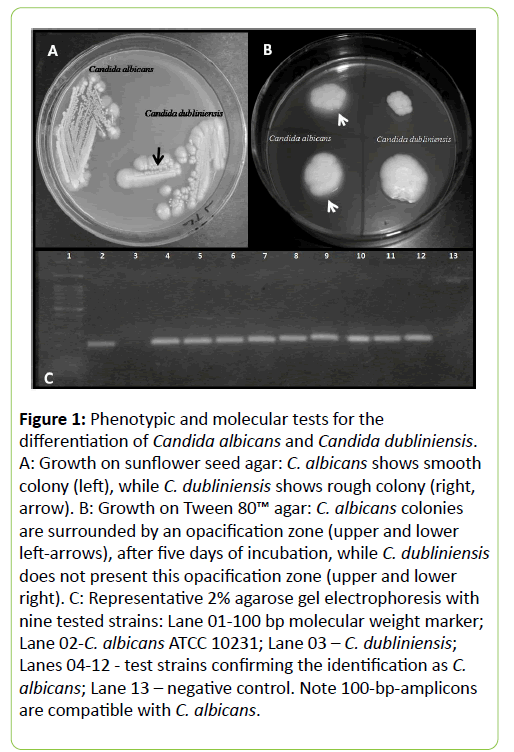
As for the phenotypic differentiation, 36 (97.3%) strains showed smooth colonies, when they were grown on sunflower seed agar, while only one (2.7%) showed rough colonies (Figure 1A). Concerning the production of esterases on Tween 80™ agar, 34 (91.9%) strains produced an opacification zone, within five days, whereas 3 (8.1%) did not produce an opacification zone, after 12 days of incubation (Figure 1B).
Figure 1: Phenotypic and molecular tests for the differentiation of Candida albicans and Candida dubliniensis. A: Growth on sunflower seed agar: C. albicans shows smooth colony (left), while C. dubliniensis shows rough colony (right, arrow). B: Growth on Tween 80™ agar: C. albicans colonies are surrounded by an opacification zone (upper and lower left-arrows), after five days of incubation, while C. dubliniensis does not present this opacification zone (upper and lower right). C: Representative 2% agarose gel electrophoresis with nine tested strains: Lane 01-100 bp molecular weight marker; Lane 02-C. albicans ATCC 10231; Lane 03 – C. dubliniensis; Lanes 04-12 - test strains confirming the identification as C. albicans; Lane 13 – negative control. Note 100-bp-amplicons are compatible with C. albicans.
Regarding the molecular identification, all 37 strains of C. albicans showed the specific 100-bp-amplicon, after PCR reaction, confirming the identification of the tested strains as C. albicans (Figure 1C).
C. dubliniensis control strains presented the expected phenotypic features for the species, i.e., germ tube production in human serum and chlamydospores on cornmeal agar, green colonies on chromogenic agar, rough colonies on sunflower seed agar (Figure 1A) and the absence of opacification zone on Tween 80™ agar, after twelve days of incubation (Figure 1B). As for the molecular identification, the specific PCR reaction with C. dubliniensis did not amplify any DNA sequence; hence, no amplicons were obtained, confirming their identity.
Discussion
C. albicans and C. dubliniensis are phylogenetically related species and share several phenotypic characteristics, often leading to misdiagnosis. The use of several phenotypic techniques has been proposed by different authors to accurately identify these species [10,17,18]. However, in many cases, the interpretation of these tests also relies on subjective descriptions, such as changes in color tones on standard chromogenic media or growth on hypertonic media [19]. C. albicans is more prevalent than C. dubliniensis as commensal and infectious agents of humans [20], and, apparently, C. albicans shows greater virulence potential in animal models, while C. dubliniensis shows greater ability to develop resistance to antifungal agents [11]. Studies with strains from animals, developed by our group, have detected the emergence of azole resistance in Candida spp. recovered from several animal species, especially C. albicans, demonstrating the importance of researches assessing this subject [12-15].
In this study, phenotypic methods were not absolutely effective for the differentiation of C. albicans and C. dubliniensis. However, the combined application of both methods (growth on sunflower seed agar and Tween 80™ agar) allowed the reliable presumptive identification of these cryptic species. Both phenotypic tests were compatible with C. albicans in 33/37 strains. Although some strains did not present the expected pattern for the species, none of the 37 tested C. albicans simultaneously showed atypical phenotypic behavior in both phenotypic tests. This association of tests seems to be an interesting tool to resolve the diagnostic confusion between yeast species, especially in small and medium routine diagnostic laboratories without access to molecular techniques. Furthermore, our findings show that the tests previously used for the identification of the strains were sufficiently accurate, since all isolates were confirmed as C. albicans by molecular biology.
Thus, our study shows that the application of classical methods in the identification of these cryptic species, including color and aspect on chromogenic agar and germ tube and chlamydospore formation, is important to identify Candida strains as C. albicans/C. dubliniensis. Moreover, the combined assessment of growth on sunflower seed agar and Tween 80™ agar improves the accuracy of phenotypic identification for the differentiation of C. albicans and C. dubliniensis.
Funding
This work was supported by grants from the National Council for Scientific and Technological Development (CNPq; Brazil; 307606/2013-9; 443167/2014-1).
Competing Interests
The authors have no conflicts of interest.
References
- Sanguinetti M, Posteraro B, Lass-Florl C (2015) Antifungal drug resistance among Candida species: mechanisms and clinical impact. Mycoses 58: 2-13.
- Sullivan DJ, Westerneng TJ, Haynes KA, Bennett DE, Coleman DC (1995) Candida dubliniensis sp. nov.: phenotypic and molecular characterization of a novel species associated with oral candidosis in HIV-infected individuals. Microbiology 141: 1507-1521.
- Bhai N, Tendolkar U, Baradkar V, Mathur M, Kulkarni M (2014) Paediatricoropharyngeal and cutaneous candidiasis with special reference to Candida dubliniensis. J Med Microbiol 63:518-521.
- Wahab AA, Taj-Aldeen SJ, Kolecka A, ElGindi M, Finkel JS, et al. (2014) High prevalence of Candida dubliniensis in lower respiratory tract secretions from cystic fibrosis patients may be related to increased adherence properties. Int J Infect Dis 24:14-19.
- Khan Z, Ahmad S, Joseph L, Chandy R (2012) Candida dubliniensis: An appraisal of its clinical significance as a bloodstream pathogen. PLoS One 7:e32952.
- McManus BA, Sullivan DJ, Moran GP, d’Enfert C, Bougnoux ME, et al. (2009) Genetic differences between avian and human isolates of Candida dubliniensis. Emerg Infect Dis 15: 1467-1470.
- Nunn MA, Schafer SM, Petrou MA, Brown JRM (2007) Environmental source of Candida dubliniensis. Emerg Infect Dis 13:747-750.
- Ahmad S, Khan Z, Asadzadeh M, Theyyathel A, Chandy R (2012) Performance comparison of phenotypic and molecular methods for detection and differentiation of Candida albicans and Candida dubliniensis. BMC Infect Dis 12:230.
- Fricke S, Fricke C, Schimmelpfennig C, Oelkrug C, Schonfelder U, et al. (2010) A real-time PCR assay for the differentiation of Candida species. J Appl Microbiol 109:1150-1158.
- Gamarra S, Mancilla E, Dudiuk C, Garcia-Effron G (2015) Candida dubliniensis and Candida albicans differentiation by colony morphotype in Sabouraud-triphenyltetrazolium agar. Rev IberoamMicol 32:126-128.
- Moran GP, Coleman DC, Sullivan DJ (2012) Candida albicans versus Candida dubliniensis: Why Is C. albicans more pathogenic? Int J Microbiol 205921.
- Sidrim JJ, Carvalho VL, Castelo-Branco DSCM, Brilhante RS, Guedes GMM, et al. (2016) Antifungal resistance and virulence among Candida spp. from captive Amazonian manatees and West Indian Manatees: potential impacts on animal and environmental health. Ecohealth 13:328-338.
- Brilhante RS, Alencar LP, Cordeiro RA, Castelo-Branco DSCM, Teixeira CE, et al. (2013) Detection of Candida species resistant to azoles in the microbiota of rheas (Rhea americana): possible implications for human and animal health. J Med Microbiol 62:889-895.
- Brilhante RS, Silva ST, Castelo-Branco DSCM, Teixeira CE, Borges LC, et al. (2015) Emergence of azole-resistant Candida albicans in small ruminants. Mycopathologia 180: 277-280.
- Castelo-Branco DSCM, Brilhante RS, Paiva MA, Teixeira CE, Caetano EP, et al. (2013) Azole-resistant Candida albicans from a wild Brazilian porcupine (Coendouprehensilis): a sign of an environmental imbalance? Med Mycol 51:555-560.
- Pfaller MA, Houston A, Coffmann S (1996) Application of CHROMagar Candida Medium™ for rapid screening of clinical specimens for Candida albicans, Candida tropicalis, Candida krusei, and Candida (Torulopsis) glabrata. Microbiology 34:58-61.
- Khan ZU, Ahmad S, Mokaddas E, Al-Sweih N, Chandy R (2005) Sunflower seed husk agar: a new medium for the differentiation of Candida dubliniensis from Candida albicans. Indian J Med Microbiol23: 182-185.
- Slifkin M (2000) Tween 80™ opacity test responses of various Candida species. J ClinMicrobiol 38: 4626-4628.
- Kiraz N, Oz Y, Aslan H, Muslumanoglu H (2014) The usefulness of DNA sequencing after extraction by Whatman FTA Filter Matrix Technology and phenotypic tests for differentiation of Candida albicans and Candida dubliniensis. Mycopathologia 177: 81-86.
- Ng KP, Kuan CS, Kaur H, Na SL, Atiya N, et al. (2015) Candida species epidemiology 2000-2013: a laboratory-based report. Trop Med Int Heal 20:1447-1453.
Open Access Journals
- Aquaculture & Veterinary Science
- Chemistry & Chemical Sciences
- Clinical Sciences
- Engineering
- General Science
- Genetics & Molecular Biology
- Health Care & Nursing
- Immunology & Microbiology
- Materials Science
- Mathematics & Physics
- Medical Sciences
- Neurology & Psychiatry
- Oncology & Cancer Science
- Pharmaceutical Sciences