Determination of Azole Antifungal Drug Resistance Mechanisms Involving Cyp51a Gene in Clinical Isolates of Aspergillus fumigatus and Aspergillus niger
Mahindran Rajendran, Tzar Mohd Nizam Khaithir and Jacinta Santhanam
DOI10.21767/2471-8521.100011
Mahindran Rajendran1*, Tzar Mohd Nizam Khaithir2 and Jacinta Santhanam1
1Biomedical Science Programme, Faculty of Allied Health Sciences, Universiti Kebangsaan Malaysia, Kuala Lumpur, Malaysia
2Department of Medical Microbiology & Immunology, Faculty of Medicine, Universiti Kebangsaan Malaysia Medical Centre, Kuala Lumpur, Malaysia
- *Corresponding Author:
- Mahindran Rajendran
Biomedical Science Programme
Faculty of Allied Health Sciences
Universiti Kebangsaan Malaysia
Jalan Raja Muda Abdul Aziz, 50300 Kuala Lumpur
Malaysia
Tel: +603-9289 7039
Fax: +603-26914304
E-mail: mahinz21@yahoo.com
Received date: October 30, 2015; Accepted date: January 21, 2016; Published date: January 28, 2016
Citation: Rajendran M, et al. Determination of Azole Antifungal Drug Resistance Mechanisms Involving Cyp51a Gene in Clinical Isolates of Aspergillus fumigatus and Aspergillus niger. Med Mycol Open Access. 2015, 2:11. doi: 10.21767/2471-8521.100011
Copyright: © 2016 Rajendran M, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Abstract
Aims: The main aim of this research is to investigate azole resistance mechanisms in A. fumigatus and A. niger which involve Cyp51A gene that encodes 14-α sterol demethylase enzyme.
Methodology and Results: Itraconazole susceptibility was determined through E-test method. A conventional PCR method was used to amplify and sequence Cyp51A gene in fungal DNA, to detect the presence of gene mutations. Real-time PCR method was applied to determine overexpression of Cyp51A gene in A. fumigatus and A. niger isolates. Susceptibility test found that 3/13 (23.1%) A. fumigatus and 7/23 (30.4%) A. niger isolates were resistant to Itraconazole, with minimum inhibitory concentrations (MICs) of 2.5 μg/ml to 3.0 μg/ml. Sequencing of A. fumigatus DNA showed presence of L98H mutation in 7/13 (53.8%) and M220 mutation in 3/13 (23%) isolates. Whereas, sequencing of A. niger DNA detected the presence of G427S mutation in 3/23 (13%) isolates. Tandem Repeat mutation was not detected in all A. fumigatus and A. niger isolates. Only M220 mutation showed significant correlation (r (13)=0.041038, p<0.05) with Itraconazole antifungal resistance in A. fumigatus isolates while L98H mutation was not involved. G427S mutation also showed correlation (r (15)=0.038434, p<0.05) with Itraconazole antifungal resistance in A. niger isolates. A higher level of Cyp51A gene expression was detected in 4/8 (50%) A. fumigatus isolates and 7/12 (58.3%) A. niger isolates. Resistant isolates more often showed higher level of Cyp51A gene expression compared to susceptible isolates; however the difference in level of expression between resistant isolates and susceptible isolates is not significant. This may be due to similar MIC values in resistant and susceptible isolates.
Conclusion, significance and impact of study: In conclusion the level of azole resistance in A. fumigatus and A. niger isolates in Malaysia is low and mutations in Cyp51A gene may contribute towards Itraconazole antifungal resistance, however other factors may also be involved.
Keywords
Aspergillus fumigatus; Aspergillus niger; Itraconazole; Cyp51A; Resistance
Introduction
Pathogenic fungi often cause serious infections to humans and other organisms. Although there are more than 500 fungal species, fewer than 100 cause disease in humans. Many cases are caused by filamentous fungi such as Aspergillus fumigatus and Aspergillus niger. Aspergillus species are widespread in the environment, growing in the soil, on plants and on decomposing organic matter [1]. These moulds are often found in outdoor and indoor air, in water, on food items and dust. Infections with A. fumigatus and A. niger cause invasive aspergillosis worldwide, resulting in high rates of mortality and morbidity in immunocompromised patients [2].
Various antifungal drugs are used as treatment for fungal infections including azole compounds. An increase in infections due to azole-resistant Aspergillus species has been observed leading to a higher case fatality rate among patients with azole-resistant invasive aspergillosis. In filamentous fungi, azole drugs inhibit ergosterol biosynthesis by targeting the enzyme 14-α sterol demethylase which is encoded by the gene Cyp51A [3]. By confirming that Cyp51A protein is the target of these antifungal agents, two molecular mechanisms of resistance to azole drugs have been described (a) azole drug resistance in A. fumigatus and A. niger seems to be mostly related to point mutations in Cyp51A gene, (b) overexpression of Cyp51A gene and (c) upregulation of efflux pumps [4-6].
Although these factors influence antifungal drug resistance but other possible mechanisms have yet to be determined. Regarding the modification of A. fumigatus and A. niger Cyp51A gene, specific mutations have been associated with susceptibility profiles whereby cross-resistance to Itraconazole has been associated with amino acid substitutions at Leucine 98 (L98H) and Methionine 220 (M220) of the target protein. It has been determined that a base change causing an amino acid substitution in Cyp51A (L98H) in combination with the duplication in tandem of a 34-bp sequence in the Cyp51A promoter, which is responsible for the increased level of Cyp51A gene expression, accounted for resistance [7].
There is minimal data on antifungal susceptibility of filamentous fungi in Malaysia and less is known of their resistance mechanisms, while Aspergillus is the most commonly isolated mold species [8]. Therefore, the aim of this study is to determine the in vitro suseptibility of Aspergillus fumigatus and Aspergillus niger clinical isolates in Malaysia towards Itraconazole and also to detect mutation or overexpression in Cyp51A gene that may contribute to antifungal drug resistance in both Aspergillus species.
Materials and Methods
Fungal strains and growth conditions
The fungal strain used in the study were (i) A. fumigatus strains ATCC 22019, ATCC 204305, reference strain F/19029; (ii) A. niger strains ATCC 6275, ATCC 90028, reference strain F/ 13295 (iii) Quality control strains ATCC 90028 and ATCC 22019. The reference strains were kindly provided by Dr. David W. Denning (University of Manchester, United Kingdom). A total of 13 clinical isolates of Aspergillus fumigatus and 23 clinical isolates of Aspergillus niger from various source were collected from the Mycology Unit, Universiti Kebangsaan Malaysia, Medical Centre and evaluated for susceptibility towards Itraconazole. The fungi were grown at room temperature on Sabouraud dextrose agar (Merck, German) and the fungus stocks were preserved in potato dextrose agar slants (Merck, German) at 4°C.
Culture conditions and antifungal susceptibility testing
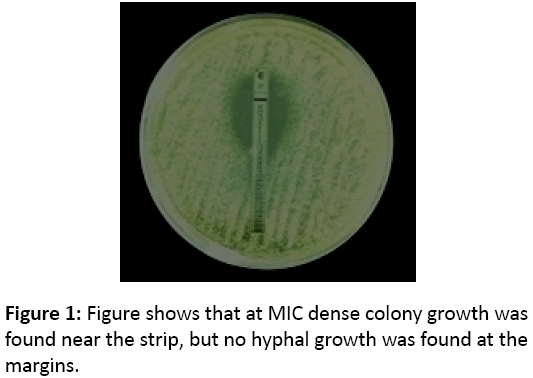
The clinical isolates fungals were cultured on PDA agar at room temperature for 48 hours to obtain the fungal inoculum at the desired colony forming unit (CFU) size. The inoculum suspension of the fungal conidia was prepared at 0.4 × 103 CFU/ml to 0.5 ×103 CFU/ml using spectrophotometer (530 nm) (OD; 0.09-0.13) as described in CLSI (formerly NCCLS) document M38-A (33) [9]. Susceptibility assay was performed by the E-test method according to the manufacturer’s instructions (AB Biodisk, Sweden). A. fumigatus and A. niger conidia were plated onto RPMI 1640 agar supplemented with 2% glucose, buffered with 0.165 M MOPS (3-(N-morpholinopropanesulfonic acid) containing L-glutamine and sodium bicarbonate and the plate was allowed to dry. E-test strips containing Itraconazole were applied, and the MIC was determined after 48 hour. The MIC was considered to be the drug concentration at which dense colony growth intersected the strip, but sparse subsurface hyphal growth at the margins was ignored (Figure 1). Candida albicans ATCC 90028 and Candida parapsilosis ATCC 22019 were used as quality control strains and all results were within the target range.
Fungal DNA extraction
Extraction of DNA from fungal cultures on PDA was performed using Dneasy Plant Mini kit, (Qiagen, Germany) according to the manufacturer’s instructions.
Fungal RNA extraction
All selected sample isolates were processed in liquid nitrogen for hyphal disruption. Extraction of RNA from fungal cultures on SDA was performed using RNeasy Plant Mini kit, (Qiagen, Germany) according to the manufacturer’s instructions.
Primers for PCR assays and sequencing
Four different primer sets for amplification of four Cyp51A gene mutations were synthesized by 1st Base, Seri Kembangan, Malaysia (Table 1). All the primers were selected based on pervious study [10]. Wild type strains A. fumigatus Cyp51A gene (GenBank accession number AF338659.1) and A. niger Cyp51A gene (GenBank accession number JF450900) were used as reference. PCRs were performed in 20 μl, with 2 μm primers, 2 μg DNA template, 0.5x Hotstar Taq Plus Master Mix (Qiagen, Germany) and 1 μl free RNase water. Thermal cycling profiles for PCR amplification were as follows: 5 min at 94°C, 45 sec at 58°C and 2 min at 72°C for first cycle, followed by 30 cycles of 30 sec at 94°C, 45 min at 98°C, extension at 72°C for 2 min and within cubation at 37°C. The final extension step is at 72°C for 10 min. The PCR products were analyzed by agarose gel electrophoresis and purified using QIAquick PCR Purification kit, (Qiagen, Germany) according to the manufacturer’s instructions for sequencing. Nucleotide sequencing analysis was performed by automated DNA sequencing. The sequence of the products was compared to the sequence of the A. fumigatus Cyp51A wild type sequence using the NCBI alignment service Align Sequence Nucleotide Blast and ClustalW tool .
| Primers | Primer Sequnce | Amplicon size (bp) | |
|---|---|---|---|
| (A. fumigatus) | CypA-L98H-S_A (F) | 5’AAAAAACCACAGTCTACCTGG 3’ | 512 |
| CypA-M220-AS_A (R) | 5’CTGATTGATGATGTCAACGTA 3’ | ||
| (A. fumigatus) | CypA-TR-S_A (F) | 5’AGCACCACTTCAGAGTTGTCTA 3’ | 100 |
| CypA-TR-AS_A (R) | 5’TGTATGGTATGCGGAACTACACCTT 3’ | ||
| (A.niger) | Ancyp51A1 (F) | 5’AACAATCTTTCTCATCAACTGGTCC 3’ | 190 |
| Ancyp51A5 (R) | 5’GATGCTTATTACAAGGTACTAGTTGG 3’ |
Table 1: A. fumigatus and A. niger Cyp51A gene primer sets. aPCR and sequencing primers: F, forward strand; R, reverse strand.
Primers for Real Time PCR and gene expression
Two primer sets for both Aspergillus species and housekeeping gene (β-Actin) were used (Table 2). The cDNA was synthesized from the isolated mRNA to DNA complementary using RT-PCR Quanti Fast SYBR Green Master Mix Real Time PCR reaction (Qiagen, Germany).
| Primers | Primer Sequnce |
|---|---|
| CypA_ F (F) | 5′-TCCTGCTCCTTAGTAGCCTGGTT -3′ |
| CypA_R (R) | 5′-GTGCTCCTTGCTTCACCTG -3′ |
| β-Actin_F (F) | 5′-ATTGCTCCTCCTGAGCGTAAATAC-3′ |
| β-Actin_R (R) | 5′-GAAGGACCGCTCTCGTCGTAC-3′ |
Table 2: Real Time PCR primer sets. aReal Time PCR primers: F, forward strand; R, reverse strand.
Real Time PCRs were performed in 25 μl, with 2 μm primers, 1 μg cDNA template (approximately70-90 mg), 2x QuantiFast SYBR Green PCR Master Mix (Qiagen, Germany) and 1 μl free RNase water. Thermal cycling profiles for Real Time PCR amplification were as follows: 3 min at 95°C, 10 sec at 95°C followed by 40 cycles and with a final extension step at 55°C for 30 sec.
Data analysis
The significance of the different mutation in Aspergillus fumigatus and Aspergillus niger isolates was determined by Chi-Square after logarithmic conversion of the values (unpaired, unequal variance). For statistical evaluation of the crossing point and relative expression variations, the data were analyzed by analysis of variance for significant differences. Statistical analysis was done with the SPSS package (version 14.0; SPSS SL., Madrid, Spain). A P value of <0.05 was considered significant.
Results
Antifungal susceptibility testing
Antifungal susceptibility data showed only 3 Aspergillus fumigatus and 7 Aspergillus niger isolates with low level resistance to Itraconazole antifungal agent (MIC: 2.5 μg/ml-3.0 μg/ml (Table 3).
| Isolates | MIC(μg/mL) E-Test ITR | Mutation | ||||||
|---|---|---|---|---|---|---|---|---|
| CYP51A gene Expression | ||||||||
| A. fumigatus | A. niger | High | Low | |||||
| L98H | M220 | TR | G427S | |||||
| A. Fumigatus | C21 | 2.0 (S) | + | - | - | - | 1.9 | - |
| C53 | 3.0 (R) | + | - | - | - | 2 | - | |
| M310 | 2.0 (S) | + | - | - | - | - | 0.4 | |
| M965 | 0.75 (S) | - | - | - | - | - | N.D | |
| M976 | 1.0 (S) | - | - | - | - | - | N.D | |
| M1420 | 1.5 (S) | - | - | - | - | - | N.D | |
| M1663 | 1.5 (S) | - | - | - | - | - | N.D | |
| M2470 | 2.0 (S) | + | - | - | - | 1 | - | |
| UZ23 | 3.0 (R) | + | + | - | - | 1 | - | |
| UZ59 | 2.0 (S) | - | - | - | - | - | N.D | |
| UZ165 | 2.0 (S) | - | + | - | - | - | 0.6 | |
| UZ291 | 2.0 (S) | + | - | - | - | - | 0.9 | |
| UZ685 | 3.0 (R) | + | + | - | - | 1 | - | |
| A. Niger | M309/12 | 3.0 (R) | - | - | - | N.D | 3.3 | - |
| M2502/12 | 1.5 (S) | - | - | - | N.D | - | 0.3 | |
| M2463/11 | 1.5 (S) | - | - | - | N.D | - | 0.1 | |
| M1587/12 | 1.0 (S) | - | - | - | N.D | - | N.D | |
| M1459/12 | 0.75 (S) | - | - | - | N.D | - | N.D | |
| M407 | 3.0 (R) | - | - | - | + | 2.2 | - | |
| M254 | 2.5 (R) | - | - | - | N.D | 1.6 | - | |
| M854 | 2.0 (S) | - | - | - | N.D | 3.3 | - | |
| M701/1 | 2.5 (R) | - | - | - | N.D | 3.4 | - | |
| M701/2 | 2.5 (R) | - | - | - | N.D | 6.9 | - | |
| M046/12 | 2.5 (R) | - | - | - | + | 1 | - | |
| M1772 | 3.0 (R) | - | - | - | + | 6.6 | - | |
| M1483 | 1.0 (S) | - | - | - | N.D | - | N.D | |
| M0200 | 2.0 (S) | - | - | - | N.D | - | N.D | |
| M167 | 2.0 (S) | - | - | - | N.D | - | N.D | |
| M054 | 1.5 (S) | - | - | - | N.D | - | N.D | |
| M166 | 2.0 (S) | - | - | - | N.D | - | N.D | |
| MM1008 | 2.0 (S) | - | - | - | N.D | - | N.D | |
| MM695 | 2.0 (S) | - | - | - | N.D | - | N.D | |
| M895 | 2.0 (S) | - | - | - | N.D | - | N.D | |
| MM1769 | 2.0 (S) | - | - | - | N.D | - | N.D | |
| M1769 | 2.0 (S) | - | - | - | N.D | - | N.D | |
| MM046/12 | 2.0 (S) | - | - | - | N.D | - | N.D | |
Table 3: In vitro susceptibilities, mutation and level of Cyp51A gene expression of 12 Aspergillus niger and 13 Aspergillus fumigatus isolates towards Itraconazole antifungal agent. a ITR; Itraconazole b MIC (EUCAST): >2.0 μg/ml; Resistant (R) ≤ 2.0 μg/ml; Susceptible (S)C N.D; Not Detected d Expression Level (High); ≥ 1.0 e Expression Level (Low); <1.0 f+Mutation present g- Mutation absent.
PCR amplification and sequence analysis of CYP51A gene
The PCR amplification showed the presence of L98H and M220 mutation using CypA-L98H-S_A and CypA-M220-AS_A set primers in Aspergillus fumigatus isolates based on the amplicon size (500 bp) and the presence of TR mutation using CypA-TR-S_A and CypA-TR-AS_A set primers were not found in both Aspergillus niger and Aspergillus fumigtus isolates (Figure 2b). The G427S mutations were only found in 3 Aspergillus niger isolates using An cyp51A1 and An cyp51A5 set primers.
Meanwhile the sequencing data showed 53.8% (7 isolates) contains L98H mutation, M220 in 23% (3 isolates) in both Aspergillus fumigtus and Aspergillus niger isolates. Azoleresistant A. fumigatus isolate F/19029 and A. niger isolate F/ 13295 served as the positive control for the detection of the L98H, M220, TR and G427S alterations in the Cyp51A gene via PCR and consecutive DNA sequence analysis. The sequence analysis of CYP51A gene in 13 A. fumigatus which has different susceptibilty (MICs) range towards Itraconazole is compared with wild type A. fumigatus (Genbank ID: AF338659.1) and showed the nucleotide changes from ‘T’ to ‘A’ at codon 364 (L98H mutation) in 7 isolates (53.8%). The nucleotide changes from ‘G’ to ‘C’ at codon 731 (M220 mutation) was found in 3 (23%) A. fumigatus isolates (Table 3). The DNA sequences of 3 A. niger isolates compared with wild type A. niger (Genbank ID: JF450900) showed the nucleotide changes from ‘G’ to ‘C’ at codon 427 indicating G427S mutation (Table 3).
Levels of Cyp51A expression by A. fumigatus and A. niger azole-resistant strains
Total of 5 Aspergillus fumigatus and 8 Aspergillus niger isolates showed overexpression of Cyp51A gene. Comparison of level of Cyp51A gene expression between resistant and susceptible isolates of Aspergillus fumigatus and Aspergillus niger respectively were analyzed statistically, however a significant differences was not found. The experiment was repeated in triplicates for both species and β-Actin gene was used as housekeeping gene. Isolate UZ685 A. fumigatus and M046 A. niger was used as calibrator.
Discussion
In this study, both Aspergillus fumigatus and Aspergillus niger showed low level of resistance to Itraconazole with MIC values between 2.5 μg/ml-3.0 μg/ml, unlike studies in United Kingdom that found 50%-70% of isolates with high level of azole resistance (MIC>8 μg/ml) [11]. The susceptibility test results clearly showed that isolates of Aspergillus fumigatus and Aspergillus niger were mostly susceptible towards Itraconazole with 72% of isolates, (n=36) with MIC<2 μg/ml. The E-test method was used based on the EUCAST method guidelines and according to Denning et al (1996) Itraconazole antifungal drug can be used as treatment fo0072 invasive Aspergillosis and gives less gives side effects compared to amphotericin B.
In determination of the resistance mechanism in Cyp51A gene towards Itraconazole, sequence analysis and level Cyp51A gene expression were studied. Primer sets were selected for Cyp51A gene amplification based on previous studies. Based on sequence analysis Aspergillus fumigatus isolates C21, C53, M310, M2470 and UZ291 showed the presence of L98H mutation. Meanwhile M220 mutation was detected in UZ165 isolate and both L98H and M220 mutation were detected in UZ23 and UZ685 isolates. The tandem repeat (TR) mutation was determined based on the amplicon size (100 bp) and this mutation was not detected as there was no increase of 34 bp in the amplicons [12].
Based on previous research [13] the cross resistance towards azole drugs is related to expression level of Cyp51A gene which is due to a Tandem Repeat 34 bp in the promoter region and amino acid changes at location 98 leucine (TRL98H). The L98H, M220 and Tandem Repeat were not detected at all in Aspergillus niger isolates and the resistance in A. niger may be due to other factors [6]. Meanwhile the resistance isolates which do not show any gene expression or L98H, M220 and TR mutation could be due to presence of other mutations such as F46Y, G89G, M172V, N248 T, D255E, L358L, E427K, C454C, L358L or efflux pump mechanism [14].
While the level of Cyp51A gene expression is higher especially in resistant A. niger isolates this increase is not significant. The presence of M220 mutation correlated significantly with resistance in A. fumigatus, however the number of resistant isolates tested were very few (3/13). The G427S mutation was detected in M046, M407 and M1772 isolates of A. niger which may indicate a significant correlation with Itraconazole antifungal drug resistance. However it was not possible to amplify the other isolates with the primer pair employed. This is because A. niger is a species complex consisting of numerous different strains [14] and several other primer pairs would have to be used in order to amplify the other isolates [15-29].
In conclusion, a very low level of resistance towards Itraconazole antifungal drug was detected in Aspergillus fumigatus and Aspergillus niger in Malaysia, which may be due to mutations in the Cyp51A gene.
Acknowledgment
We thank Dr. David W. Denning (University of Manchester, United Kingdom) for providing the reference strains F/19029 (A. fumigatus) and F/13295 (A. niger).
References
- Latgé JP (1999) Aspergillus fumigatus and aspergillosis. Clin Microbiol Rev 12: 310-350.
- Hawksworth DL (2001) The magnitude of fungal diversity: the 1.5 million species estimate revisited. Mycol Res 105: 1422-1432.
- Brookman JL, Denning DW (2000) Molecular genetics in Aspergillus fumigatus. Curr Opin Microbiol 3: 468-474.
- Howard SJ, Arendrup MC (2011) Acquired antifungal drug resistance in Aspergillus fumigatus: epidemiology and detection. Med Mycol 49 Suppl 1: S90-95.
- Snelders E, van der Lee HA, Kuijpers J, Rijs AJ, Varga J, et al. (2008) Emergence of azole resistance in Aspergillus fumigatus and spread of a single resistance mechanism. PLoS Med 5: e219.
- Denning DW (1996) Diagnosis and management of invasive aspergillosis. Curr Clin Top Infect Dis 16: 277-299.
- Tzar MN, Suhaila B, Shamsul AS, Azizah M (2013) Epidemiology of fungal infections at an infectious disease reference centre in Malaysia. International Medical Journal Malaysia 12: 39-42.
- Espinel-Ingroff A, Diekema DJ, Fothergill A, Johnson E, Pelaez T, et al. (2010) Wild-type MIC distributions and epidemiological cutoff values for the triazoles and six Aspergillus spp. for the CLSI broth microdilution method (M38-A2 document). J Clin Microbiol 48: 3251-3257.
- Mellado E, Diaz-Guerra TM, Cuenca-Estrella M, Rodriguez-Tudela JL (2001) Identification of two different 14-_ sterol demethylase-related genes (cyp51A and cyp51B) in Aspergillus fumigatus and other Aspergillus species. J Clin Microbiol 39: 2431–2438.
- Denning DW, Park S, Lass-Florl C, Fraczek MG, Kirwan M, et al. (2011) High-frequency triazole resistance found In nonculturable Aspergillus fumigatus from lungs of patients with chronic fungal disease. Clin Infect Dis 52: 1123-1129.
- Garcia-Effron G, Dilger A, Alcazar-Fuoli L, Park S, Mellado E, et al. (2008) Rapid detection of triazole antifungal resistance in Aspergillus fumigatus. J Clin Microbiol 46: 1200-1206.
- Denning DW, Venkateswarlu K, Oakley KL, Anderson MJ, Manning NJ, et al. (1997) Itraconazole resistance in Aspergillus fumigatus. Antimicrob Agents Chemother 41: 1364-1368.
- Bueid A, Howard SJ, Moore CB, Richardson MD, Harrison E, et al. (2010) Azole antifungal resistance in Aspergillus fumigatus: 2008 and 2009. J Antimicrob Chemother 65: 2116-2118.
- Howard SJ, Elizabeth H, Paul B, Janos V, David WD (2011) Cryptic Species and Azole Resistance inthe Aspergillus niger Complex. Antimicrob Agents Chemother 55: 4802-4809.
- Denning DW (1998) Invasive aspergillosis. Clin Infect Dis 26: 781-803.
- Howard SJ, Cerar D, Anderson MJ, Albarrag A, Fisher MC, et al. (2009) Frequency and evolution of Azole resistance in Aspergillus fumigatus associated with treatment failure. Emerg Infect Dis 15: 1068-1076.
- Howard SJ, Webster I, Moore CB, Gardiner RE, Park S, et al. (2006) Multi-azole resistance in Aspergillus fumigatus. Int J Antimicrob Agents 28: 450-453.
- Mellado E, Garcia-Effron G, Alcazar-Fuoli L, Cuenca-Estrella M, Rodriguez-Tudela JL (2004) Substitutions at methionine 220 in the 14-alpha sterol demethylase (Cyp51A) of Aspergillus fumigatus are responsible for resistance in vitro to azole antifungal drugs. Antimicrob Agents Chemother 48: 2747–2750.
- Mellado E, Garcia-Effron G, Buitrago MJ, Alcazar-Fuoli L, Cuenca-Estrella M, et al. (2005) Targeted gene disruption of the14-alpha sterol demethylase (Cyp51A) in Aspergillus fumigatus and its role in azole drug susceptibility. Antimicrob Agents Chemother 49: 2536-2538.
- Mellado E, Garcia-Effron G, Alcázar-Fuoli L, Melchers WJ, Verweij PE, et al. (2007) A new Aspergillus fumigatus resistance mechanism conferring in vitro cross-resistance to azole antifungals involves a combination of cyp51A alterations. Antimicrob Agents Chemother 51: 1897-1904.
- National Committee for Clinical Laboratory Standards (2002) Reference method for broth dilution antifungal susceptibility testing of filamentous fungi: approved standard M38-A. Na.
- Perlin DS (2009) Antifungal drug resistance: do molecular methods provide a way forward? Curr Opin Infect Dis 22: 568-573.
- Pfaller MA, Messer SA, Hollis RJ, Jones RN (2002) Antifungal activities of posaconazole, ravuconazole, and voriconazole compared tothose of itraconazole and amphotericin B against 239 clinical isolates of Aspergillus spp. and other filamentous fungi: report from SENTRY Antimicrobial Surveillance Program, 2000. Antimicrob Agents Chemother 46: 1032-1037.
- Pfaller MA, Diekema DJ, Ghannoum MA, Rex JH, Alexander BD, et al. (2009) Wild-type MIC distribution and epidemiological cut off values for Aspergillus fumigatus and three triazoles as determined bythe Clinical and Laboratory Standards Institute broth microdilution methods. J Clin Microbiol 47: 3142-3146.
- Pfaller M, Boyken L, Hollis R, Kroeger J, Messer S, et al. (2011) Use of epidemiological cutoff values to examine 9-year trends in susceptibility of Aspergillus species to the triazoles. J Clin Microbiol 49: 586-590.
- Rodriguez-Tudela JL, Alcazar-Fuoli L, Mellado E, Alastruey-Izquierdo A, Monzon A, et al. (2008) Epidemiological cutoffs and cross-resistance to azole drugs in Aspergillus fumigatus. Antimicrob Agents Chemother 52: 2468-2472.
- Verweij PE, Mellado E, Melchers WJ (2007) Multiple-triazole-resistant aspergillosis. N Engl J Med 356: 1481-1483.
- Verweij PE, Howard SJ, Melchers WJ, Denning DW (2009) Azole-resistance in Aspergillus: proposed nomenclature and breakpoints. Drug Resist Updat 12: 141-147.
- Verweij PE, Snelders E, Kema GH, Mellado E, Melchers WJ (2009) Azole resistance in Aspergillus fumigatus: a side-effect of environmental fungicide use? Lancet Infect Dis 9: 789-795.
Open Access Journals
- Aquaculture & Veterinary Science
- Chemistry & Chemical Sciences
- Clinical Sciences
- Engineering
- General Science
- Genetics & Molecular Biology
- Health Care & Nursing
- Immunology & Microbiology
- Materials Science
- Mathematics & Physics
- Medical Sciences
- Neurology & Psychiatry
- Oncology & Cancer Science
- Pharmaceutical Sciences