Molecular Identification and Antifungal Susceptibility of Candida parapsilosis sensu stricto, Candida orthopsilosis, and Candida metapsilosis in Sousse Region, Tunisia
Ben Hadj HassineAhmed,Ben AbdeljelilJihene,Ben Said Moncef,Boukadida Jalel and Saghrouni Fatma
DOI10.21767/2471-8521.100008
Ben Hadj Hassine Ahmed*, Ben Abdeljelil Jihene, Ben Said Moncef, Boukadida Jalel and Saghrouni Fatma
UR 12SP34 Research Unit, minutesistry of Public Health, Tunis, Tunisia
- *Corresponding Author:
- Ben Hadj Hassine Ahmed
UR 12SP34 Research Unit
Ministry of Public Health
Tunis, Tunisia
Tel: +216-40-143-343
E-mail: benhadjhassine.ahmed@yahoo.fr
Received date: October 14, 2015; Accepted date: December 02, 2015; Published date: December 09, 2015
Citation: Ahmed BHH, Jihene BA, Moncef BS, et al. Molecular Identification and Antifungal Susceptibility of Candida parapsilosis sensu stricto, Candida orthopsilosis, and Candida metapsilosis in Sousse Region, Tunisia. Med Mycol Open Access. 2015, 1:8. doi: 10.21767/2471-8521.100008
Copyright: © 2016 Ahmed BHH, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Abstract
Candida parapsilosis was recently reclassified into three distinct species, C. parapsilosis sensu stricto, C. orthopsilosis, and C. metapsilosis. The aim of this study was to assess the prevalence and the antifungal susceptibility of these species among clinical isolates previously identified by phenotypical methods as C. parapsilosis in the laboratory of parasitology of Farhat Hached Hospital (Sousse, Tunisia). Ninety-six clinical isolates were identified by using a multiplex PCR targeting the gene encoding the ribosomal protein SO 40 (RPSO) and a PCR targeting the ITS region. Ninety one (94.8%) isolates were identified as C. parapsilosis sensu stricto, 3 (3.1%) as C. metapsilosis, and 2 (2.1%) as C. orthopsilosis based on PCRITS. These results were concordant with those achieved by Multiplex PCR-RPSO but the prevalence of C. orthopsilosis and C. metapsilosis seems to be very low in the region. Seventeen of the Candida isolates, C. parapsilosis sensu stricto (12), C. metapsilosis (3) and C. orthopsilosis (2) were assayed by ATB Fungus 3® to assess their susceptibility to Flucytosine, Amphotericin B, Fluconazole, Itraconazole and Voriconazole and by E-test® to assess their susceptibility to Caspofungin. All the isolates were susceptible to the antifungals tested.
Keywords
Candida parapsilosis; Candida metapsilosis; Molecular identification; Antifungal susceptibility; Tunisia
Introduction
Candida parapsilosis is an emergent pathogen and has become the second or the third most common Candida species causing invasive and superficial candidiasis [1,2]. The molecular investigations of C. parapsilosis have led to the sequencing of its full genome, which showed high heterogeneity between strains otherwise phenotypically indistinguishable [3,4]. As a result, it was established that C. parapsilosis is a complex of three genetically distinct groups: group I, group II and group III [5,6]. Detailed molecular analyses have however, recently recognized C. parapsilosis groups as separate species named C. parapsilosis, C. orthopsilosis and C. metapsilosis respectively [7]. The differentiation of the three species of C. parapsilosis complex is clinically relevant because of their virulence and their susceptibility to antifungal agents. On the other hand, the prevalence of the species varies according to the regions [8-12]. A study conducted between 1995 and 2009 at the Farhat Hached teaching Hospital of Sousse, east-central Tunisia, revealed that C. parapsilosis sensu lato was the second most-frequent Candida species isolated from patients with candidemia, and that its incidence has dramatically increased over the study period [13]. The aim of the present study was to assess the frequency of each of the three species of C. parapsilosis complex in our region by using two PCR techniques: a PCR targeting the gene encoding the ribosomal protein SO 40 (RPSO) and a PCR targeting the ITS region, followed by the sequencing of the PCR product [14,15]. Additionally, the susceptibility of clinical isolates to antifungals by using ATB Fungus 3® test and E-test® was evaluated.
Materials and Methods
Strains: A total of 96 Candida strains were isolated from patients admitted to different care units of Farhat Hached Hospital of Sousse, Tunisia, or from outdoor patients referred to the laboratory of parasitology-mycology of the same hospital between 2007 and 2013. Clinical sources of the 96 isolates are as listed in Table 1. All isolates were identified initially as C. parapsilosis sensu lato on the basis of morphological and biochemical criteria. Reference strains ATCC22019 and CECT13009 were used as controls for C. parapsilosis, while reference strains CECT13011 and CECT13010 were used as controls for C. orthopsilosis and C. metapsilosis, respectively. Isolates were kept at -80°C for long-term storage and prior to DNA extraction, isolates were grown in YPD (yeast peptone dextrose) medium at 35°C.
| Site | Number (%) |
|---|---|
| Fingers tinea unguium | 48 (50) |
| Toes tinea unguium | 9 (9.37) |
| Blood | 8 (8.33) |
| External otitis | 7 (7.29) |
| Umbilical venous catheter | 6 (6.25) |
| Inter toes intertrigo | 5 (5.20) |
| Nose | 2 (2.08) |
| Vagina | 2 (2.08) |
| Central catheter | 1 (1.04) |
| Hair | 1 (1.04) |
| Sputum | 1 (1.04) |
| Interdigital intertrigo | 1 (1.04) |
| Mouth | 1 (1.04) |
| Bronchial aspiration | 1 (1.04) |
| Tinea cruris | 1 (1.04) |
| Cerebrospinal fluid | 1 (1.04) |
| Endotracheal tube | 1 (1.04) |
| Total | 96 (100.00) |
Table 1: Distribution of tested clinical strains according to the site of isolation.
DNA extraction: Yeast DNA was extracted as described by Tavanti et al. (2007) [16]. Briefly, yeasts grown in YPD medium at 35°C for 24 hours were harvested by centrifugation at 1500 rpm for 5 minutes, and lysed by vortexing for 5 minutes with 0.3 gr of glass beads (diameter 0.45 mm to 0.52 mm; Sigma-Aldrich, USA), 200 μl of lysis buffer (100 mmol/L Tris-Hcl, pH 8.0, 2% Triton X100 [vol/vol] and 1 mmol/L EDTA) and 200 μl of 1:1 (vol/ vol) phenol-chloroform. After vortexing for 3 minutes, 200 μl of the extraction buffer (10 mmol/L Tris-HCl, pH 8.0, 1 mmol/L EDTA) were added to the lysate and the mixture centrifuged at 5000 rpm for 10 minutes. The aqueous phase was afterwards transferred to a new vial and the DNA precipitated by adding 1 ml of ethanol. After a 10 minutes centrifugation at 14000 rpm, the pellet was harvested and resuspended in 400 μl of extraction buffer with 5 μl of proteinase K (20 mg/ml) (Sigma-Aldirish, USA). The mixture was then incubated for 1 hour at 65°C and for 30 minutes at 72°C, and the DNA precipitated by the addition of 1 ml of isopropanol and 10 μl of 4 mol/L Ammonium acetate. After a 10 minutes centrifugation at 14000 rpm, the harvested DNA pellet was dried and redissolved in 50 μl of extraction buffer and cryopreserved at -20°C until use. Prior to the amplification procedure DNA concentrations and A 260 nm/A280 nm ratios were determined by means of the automated NanoDrop® ND.1000 spectrophotometer (BioRad, France). DNA was considered pure when the ratio A260nm/A280nm was between 1.8 and 2.0 one unit of optical density at 260 nm corresponding to 50 μg/ml.
Identification of isolates: Phenotypic identification: Initially, isolates were identified as C. parapsilosis on the basis of the following criteria: (i) characterization of colonies on chromogenic CAN2 medium (bioMérieux, France), (ii) microscopic features of colonies grown on PCB medium (BioRad, France) and (iii) sugar assimilation profiles with the Auxacolor 2® (BioRad, France) and ID32C (bioMérieux, France) kits.
Molecular identification: For this purpose we used two PCR methods.
PCR-RPSO: this method was carried out as described by Del Pilar Vercher et al. [14]. It is based on the amplification of the RPSO gene intron, by using 3 primers (presented in Table 2) that generate amplicons of 100 bp, 150 bp and 200 bp with C. parapsilosis, C. orthopsilosis and C. metapsilosis DNA extracts respectively. In the initial application reported by Del Pilar et al., the three primers were used separately in 3 PCR runs for each isolate [14]. In our own work, the technique was optimized in a way so that the primers were used simultaneously in a single PCR multiplex run. The PCR assay was performed in a total reaction volume of 25 μl containing 2.5 μl of 5X green Go Taq flexi buffer (Promega), 0.4 μmol/L of each primer, 400 μmol/L of 10 mmol/L dNTPs Mix (Promega), 2 mmol/L of 25 mmol/L MgCl2 (Promega), 1.25 U of Go Taq Hot start polymerase (Promega), 11.75 μl of nuclease free water (Promega) and 100 ng of genomic DNA.
| Species | Primer name | Sequence | Amplicon size (bp) |
|---|---|---|---|
| C. parapsilosis | CP1 | 5’-AGGGATTGCCAATATGCCCA-3’ | 100 |
| CP2 | 5’-GTGACATTGTGTAGATCCTTG G-3’ | ||
| C. orthopsilosis | CO1 | 5’TTTCAATATGCCTAGAGCCACATTGTGAATC-3’ | 150 |
| CO2 | 5’-GCATTAGTTAGTATCGTCTTTTATTAAATA3’ | ||
| C. metapsilosis | CM1 | 5’-AATAGAGGAGATGTTTTATTTGAATTC-3’ | 200 |
| CM2 | 5’-GCAGAATCCGTAAGAACTGGGG-3’ |
Table 2: Characteristics of primers used for PCR-RPSO.
The amplification conditions were as follows: a 3 minutes initial denaturation step at 95°C, followed by 45 cycles at 94°C for 30 s (denaturation), 63°C for 30 s (annealing), and 72°C for 54 s (extension) and a final extension step of 10 minutes at 72°C. PCR was performed in a thermal cycler Perkin Elmer GeneAmp PCR System 2400.
PCR-ITS: the PCR-ITS was performed by using the primers ITS1 (5’-CCGTAGGTGAACCTGCGG-3’) and ITS4 (5’-TCCTCCGCTTATTGATATGC-3’) described by White et al. [15]. These primers amplify a 500 bp product that encompasses the entire ITS1, the 5.8s and the ITS2 gene regions. The PCR assay was carried out according to the procedure described by Pryce et al. (2006), in a total reaction volume of 50μl [17]. The PCR mixture contained 5 μl of 5x PCR buffer (5X green Go Taq flexi Buffer, Promega); 3 μl of 25 mmol/L MgCl2; 3 μl of 10 μmol/L of each primer; 1 μl of 10 mmol/L of each dNTP (dNTP Mix, Promega); 2.5 U of Go Taq Hot start polymerase (Promega); and 29.5 μl of sterile distilled water (nuclease free water, Promega). The PCR was performed in a thermal cycler (Perkin Elmer GeneAmp PCR System 2400) as described by Turenne et al., with the following program: 94°C for 10 minutes (initial denaturation) followed by 94°C for 55 s (denaturation), 55°C for 60s (annealing), 72°C for 60s (extension) for 30 cycles, followed by 72°C for 10 minutes (final extension) [18].
Sequencing of the PCR-ITS products: All 96 PCR-ITS amplified products were sequenced by using the DNA Engine Tetrad 2 Peltier Thermal Cycler (BioRad, France) and the Cycle Sequencing ABI Dye Ter minutesator® kit (Applied Biosystems) according to the manufacturer’s instructions. The amplified products were sequenced in a forward and reverse direction using ITS1 and ITS4 primer, respectively. The fluorescent fragments were purified with the Big Dye X Terminator Purification Kit (Applied Biosystems), then sequenced by means of the ABI 3730xl DNA Analyzer sequencer (96 capillary type). The electrophoregrams were visualized by the Chromas Lite version 2.1.1 and the nucleotide sequences alignments performed with the (BLASTN 2.2.29+) BLAST for species identification.
Detection of PCR products: For both PCR products, the amplified products were detected by gel electrophoresis using a 2% agarose gel (UltraPure Agarose®, Invitrogen) in 0.5xTris-Acetate- EDTA buffer (UltraPure® 10X TAE buffer, Invitrogen), stained with 0.5 μg/ml ethidium bromide. The same buffer without ethidium bromide was used as electrophoresis buffer. A total volume of 5 μl of 100 bp ladder (Promega) was run in parallel with sample DNAs for PCR product band sizing. Electrophoresis conditions were 110v for 60 minutes in 0.5x TAE buffer. DNA bands were visualized by UV transillumination, photographed with the Molecular Imager® Gel Doc™ XR+ Imaging system and analyzed by using the Quantity One® software (BioRad, France).
Antifungal susceptibility testing: In order to evaluate the in vitro susceptibility profile of the clinical isolates C. parapsilosis complex strains to Flucytosine (Fc), Amphotericin B (Amb), Fluconazole (Flc), Itraconazole (Itc) and Voriconazole (VC) antifungals, 12 C. parapsilosis sensu stricto, 3 C. metapsilosis and 2 C. orthopsilosis were tested by using the ATB Fungus® kit (bioMérieux, France). The antifungals test was conducted according to the manufacturer's instructions, and reading taken after 24 hours. In addition, 17 Candida strains were tested against caspofungin (CS) by using the E-test® strips (BioMérieux, France). The test was performed by incubation of colonies of Candida isolates on agar plates containing 15 ml of casitone agar (BioRad, France). Plates were incubated at 35°C and read after 24 hours. For both ATB Fungus® and E-test®, the minimum inhibitory concentrations (CMIs) were determined for all antifungals (except FC) and interpreted according to the Clinical and Laboratory Standards Institute (CLSI) recommendations.
Results
PCR: All tested isolates were successfully amplified using both PCR-RPSO and PCR-ITS.
Out of 96 C. parapsilosis clinical isolates tested with multiplex PCR-RPSO, 91 (94.8%) gave the single 100 bp band and were assigned to the most common C. parapsilosis species; 3 (3.1%) gave 2 bands of 100 bp and 150 bp respectively and were identified as C. metapsilosis and 2 (2.1%) gave 2 bands of 100 bp and 200 bp respectively, and were identified as C. orthopsilosis.
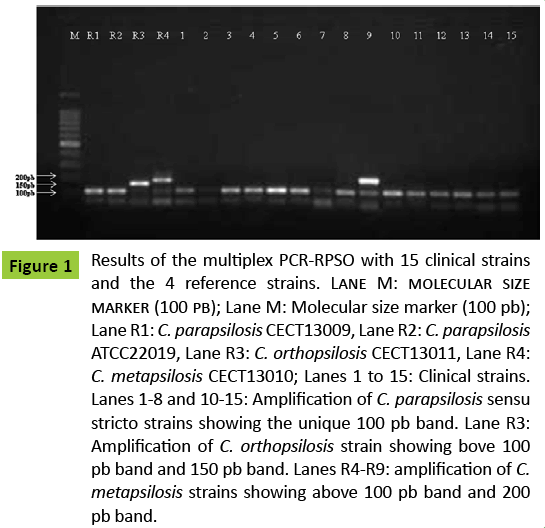
An example of the electrophoregram given by some clinical strains and by the reference strains is shown in Figure 1.
Figure 1: Results of the multiplex PCR-RPSO with 15 clinical strains and the 4 reference strains. Lane M: molecular size marker (100 pb); Lane M: Molecular size marker (100 pb); Lane R1: C. parapsilosis CECT13009, Lane R2: C. parapsilosis ATCC22019, Lane R3: C. orthopsilosis CECT13011, Lane R4: C. metapsilosis CECT13010; Lanes 1 to 15: Clinical strains. Lanes 1-8 and 10-15: Amplification of C. parapsilosis sensu stricto strains showing the unique 100 pb band. Lane R3: Amplification of C. orthopsilosis strain showing bove 100 pb band and 150 pb band. Lanes R4-R9: amplification of C. metapsilosis strains showing above 100 pb band and 200 pb band.
Out of the 3 C. metapsilosis isolates, two were collected from toes’ onychomycosis and one from a venous umbilical catheter.
Both C. orthopsilosis isolates were collected from fingers’ onychomycosis. C. parapsilosis was isolated from all types of sites and/or lesions, namely: fingers tinea unguium (46), blood (8), toes tinea unguium (7), external otitis (7), umbilical venous catheter (5), inter toes intertrigo (5), nose (2), vagina (2), central catheter (1), hair (1), sputum (1), intertrigo (1), mouth (1), bronchial aspiration (1), tinea cruris (1), cerebrospinal fluid (1), endotracheal tube (1).
All of the 96 C. parapsilosis sensu lato clinical strains gave the expected 500 bp band in PCR-ITS. In order to confirm their characterization at the species level, the ITS1/ITS2 region and the 5.8 rRNA gene previously amplified by PCR-ITS, were sequenced.
After analysis of forward and reverse sequences by nucleotide BLAST search, 91 isolates showed 96-99% similarity with the C. parapsilosis sequence, 3 isolates showed 99% similarity with the C. metapsilosis sequence, and 2 isolates showed 99% similarity with the C. orthopsilosis sequence.
Thus, according to PCR-RPSO and DNA sequencing of ITS region, 91 isolates were identified as C. parapsilosis sensu stricto; three isolates were identified as C. metapsilosis and two isolates identified as C. orthopsilosis. No discordant results were observed between the two techniques.
Antifungal susceptibility: The 17 tested strains were susceptible to all antifungal agents. MICs of AMB, FLC, ITC and VC as determined by ATB Fungus® were <0.5 μg/ml, <1 μg/ml, <0.125 μg/ml and <0.125 μg/ml, respectively for all 3 species and all isolates.
MICs of CS as determined by E-test® were 0.047 μg/ml and 0.25 μg/ml for the first and second isolates of C. orthopsilosis, respectively; and 0.064 μg/ml for all 3 isolates of C. metapsilosis. MICs ranged between 0.016 μg/ml and 0.19 μg/ml for the 91 C. parapsilosis sensu stricto isolates.
Discussion
Many molecular methods have been adapted to the identification at the species level of C. parapsilosis clinical isolates, and among them are PCR-RPSO and PCR-ITS, which are the most widely used [14,15]. Multiplex PCR-RPSO is highly recommended for the molecular identification of the three cryptic species owing to its simplicity, rapidity and cost, especially in laboratories with low income. Thus, in the present study, molecular analysis of C. parapsilosis isolates led to the separation of the species into three groups further reclassified as separate species, C. parapsilosis sensu stricto, C. orthopsilosis and C. metapsilosis, respectively [7]. But the three species were phenotypically indistinguishable, in spite of the fact that recent studies implicated the three recently described species in human diseases, including bloodstream infection [11,12,19-22].
In PCR-RPSO, which targets the gene encoding the SO40 S ribosomal protein, the size of the amplified fragment is different according to the species, and this allows a precise identification of each of the three species of the complex. However, isolates were tested in three separate runs, each using one different primers couple [14]. In contrast, in the present study, a PCRRPSO method was developed where the three primers were used simultaneously in the same run, allowing a much more rapid and less costly identification procedure. The RPSO gene was successfully amplified in all 96 clinical isolates, as well as in the four reference strains tested. According to the patterns obtained, 91 (94.8%) isolates were identified as C. parapsilosis sensu stricto, three (3.1%) as C. metapsilosis and two (2.1%) as C. orthopsilosis. For a further confirmation of these findings, all the clinical isolates were subjected to PCR-ITS and the amplification products submitted for sequencing. It is worth noting that the PCR-ITS which targets the 5.8s ribosomal gene together with the ITS1 and ITS2 flanking regions and the sequencing of the amplified products are considered as the gold standard in the distinction between the three species of C. parapsilosis complex [15,18,23]. Results of this study showed a very high concordance between PCR-RPSO and sequences of PCR-ITS amplified products. Nevertheless, PCR-RPSO was much more adapted to the hospital setting owing to its advantages in terms of simplicity, rapidity and cost especially when used in the multiplex variant we developed in the present work. Regarding the frequency of each of the three clinical species among the isolates investigated in our study, present findings are in accordance with nearly all previous reports where the prevalence of C. orthopsilosis and C. metapsilosis is less than 10% even though differences between series and regions have been described [7,8,11,12,16,19- 21]. The much higher frequency of the isolation from clinical specimens of C. parapsilosis sensu stricto than C. orthopsilosis and C. metapsilosis has been attributed, among other factors, to its easier transmission from person to person and to its better adaptation to human commensal environment than its two relatives [5,6]. We didn't observe any resistance to all antifungals tested in our 17 isolates. MICs of AMB, FLC, ITC and VC were low for all isolates of the three species. This finding is in accordance with most of previous reports [24,25]. However, the low number of isolates tested in our study makes it difficult to determine any significant conclusion especially concerning caspofungin. Indeed, C. parapsilosis sensu stricto and C. orthopsilosis isolates have been associated with lower susceptibility to and higher MICs of CS as compared to C. metapsilosis and other Candida species and MICs of micafungin have been reported to be higher towards C. parapsilosis sensu stricto when compared to both C. orthopsilosis and C. metapsilosis isolates [23]. Several recent studies highlighted the clinical relevance of both C. orthopsilosis and C. metapsilosis. C. orthopsilosis has been recovered from blood, indwelling catheters, urine, lungs, skin and genital apparatus [8,11,12,19,22]; and C. metapsilosis was reported to have caused up to 6.9% of all C. parapsilosis sensu lato fungemia cases in Spain [20]. Out of the five non C. parapsilosis sensu stricto isolates identified in this study, four were collected from superficial sites (nails). The strain identified as C. metapsilosis from an umbilical catheter may indicate the invasiveness of the strain, with a further diffusion through blood. Even though none can be considered to be resistant to CS, the relatively high MICs shown by 17 of the isolates is somewhat in agreement with the findings mentioned earlier. Concerning the commonly used azoles, C. parapsilosis sensu stricto and to a lesser extent C. metapsilosis isolates have been shown to be less sensitive to FLC. Resistance to FC which has been described among C. parapsilosis isolates is unlikely to be a major concern when considering therapeutic use of this drug in candidiasis [23]. These differences in the activity of antifungals could have clinical relevance and affect the therapeutic choices. They emphasize the need for accurate species identification and antifungal susceptibility testing of C. parapsilosis strains isolated from patients with invasive candidiasis. However, molecular methods may not be adapted to the identification of isolates in routine; so the development of new tests for phenotypical identification is highly needed. Recently, several studies have reported the efficiency of MALDITOF MS for fast and accurate identification of C. parapsilosis sensu stricto, C. metapsilosis and C. orthopsilosis [26-28].
Conflict of Interest
The author has no conflict of interest.
References
- Trofa D, Gácser A, Nosanchuk JD (2008) Candida parapsilosis, an emerging fungal pathogen. Clin Microbiol Rev 21: 606-625.
- van Asbeck EC, Clemons KV, Stevens DA (2009) Candida parapsilosis: a review of its epidemiology, pathogenesis, clinical aspects, typing and antimicrobial susceptibility. Crit Rev Microbiol 35: 283-309.
- Lott TJ, Kuykendall RJ, Welbel SF, Pramanik A, Lasker BA (1993) Genomic heterogeneity in the yeast Candida parapsilosis. Curr Genet 23: 463-467.
- Lehmann PF, Lin D, Lasker BA (1992) Genotypic identification and characterization of species and strains within the genus Candida by using random amplified polymorphic DNA. J Clin Microbiol 30: 3249-3254.
- Lin D, Wu LC, Rinaldi MG, Lehmann PF (1995) Three distinct genotypes within Candida parapsilosis from clinical sources. J Clin Microbiol 33: 1815-1821.
- Roy B, Meyer SA (1998) Confirmation of the distinct genotype groups within the form species Candida parapsilosis. J Clin Microbiol 36: 216-218.
- Tavanti A, Davidson AD, Gow NA, Maiden MC, Odds FC (2005) Candida orthopsilosis and Candida metapsilosis spp. nov. to replace Candida parapsilosis groups II and III. J Clin Microbiol 43: 284-292.
- Lockhart SR, Messer SA, Pfaller MA, Diekema DJ (2008) Geographic distribution and antifungal susceptibility of the newly described species Candida orthopsilosis and Candida metapsilosis in comparison to the closely related species Candida parapsilosis. J Clin Microbiol 46: 2659-2664.
- Gácser A, Schäfer W, Nosanchuk JS, Salomon S, Nosanchuk JD (2007) Virulence of Candida parapsilosis, Candida orthopsilosis, and Candida metapsilosis in reconstituted human tissue models. Fungal Genet Biol 44: 1336-1341.
- Németh T, Tóth A, Szenzenstein J, Horváth P, Nosanchuk JD, et al. (2013) Characterization of virulence properties in the C. parapsilosissensulato species. PLoS One 8: e68704.
- Garcia-Effron G, Canton E, Pemán J, Dilger A, Romá E, et al. (2012) Epidemiology and echinocandin susceptibility of Candida parapsilosissensulato species isolated from bloodstream infections at a Spanish university hospital. J Antimicrob Chemother 67: 2739-2748.
- Trabasso P, Matsuzawa T, Fagnani R, Muraosa Y, Tominaga K, et al. (2015) Isolation and drug susceptibility of Candida parapsilosissensulato and other species of C. parapsilosis complex from patients with blood stream infections and proposal of a novel LAMP identification method for the species. Mycopathologia 179: 53-62.
- Saghrouni F, Bougmiza I, Ben Abdeljelil J, Yacoub A, Khammari I, et al. (2011) Epidemiological trends in invasive candidiasis: results from 15-year study in Sousse region, Tunisia. J Myc Med 21: 123-129.
- Del PilarVercher M, GarcíaMartínez JM, Cantón E, Pemán J, Gómez García MM, et al. (2011) Differentiation of Candida parapsilosis, C. orthopsilosis, and C. metapsilosis by specific PCR amplification of the RPS0 intron. Int J Med Microbiol 301: 531-535.
- White TJ, Bruns T, Lee S, Taylor J (1990) Amplification and direct sequencing fungical ribosomal RNA genes for phyllogenitics. PCR protocols: a guide to methods and applications.
- Innis MA, Gelfand DH, Sninsky JJ, White TJ. Academic Press, Inc.,San Diego, California 92101, USA, pp. 315-325.
- Tavanti A, Hensgens LA, Ghelardi E, Campa M, Senesi S (2007) Genotyping of Candida orthopsilosis clinical isolates by amplification fragment length polymorphism reveals genetic diversity among independent isolates and strain maintenance within patients. J Clin Microbiol 45: 1455-1462.
- Pryce TM, Palladino S, Price DM Gardam DJ, Campbell PB, et al. (2006) Rapid identification of fungal pathogens in BacT/ALERT, BACTEC, and BBL MGIT media using polymerase chain reaction and DNA sequencing of the internal transcribed spacer regions. Diagn Microbiol Infect Dis 54: 289-297.
- Turenne CY, Sanche SE, Hoban DJ, Karlowsky JA, Kabani AM (1999) Rapid identification of fungi by using the ITS2 genetic region and an automated fluorescent capillary electrophoresis system. J Clin Microbiol 37: 1846-1851.
- González GM, Treviño-Rangel Rde J, Palma-Nicolás JP, Martínez C, González JG, et al. (2013) Species distribution and antifungal susceptibility of bloodstream fungal isolates in paediatric patients in Mexico: a nationwide surveillance study. J Antimicrob Chemother 68: 2847-2851.
- Gomez-Lopez A, Alastruey-Izquierdo A, Rodriguez D , Almirante B, Pahissa A, et al. (2008) Prevalence and susceptibility profile of Candida metapsilosis and Candida orthopsilosis: results from population-based surveillance of candidemia in Spain. Antimicrob Agents Chemother 52: 1506-1509.
- Chen CY, Sheng WH, Huang SY, Chou WC, Yao M, et al. (2015) Clinical characteristics and treatment outcomes of patients with candidaemia due to Candida parapsilosissensulato species at a medical centre in Taiwan, 2000-12. J Antimicrob Chemother 70: 1531-1538.
- Zhu Y, Shan Y, Fan S, Li J, Liu X (2015) Candida parapsilosissensustricto and the closely related species Candida orthopsilosis and Candida metapsilosis in vulvovaginal candidiasis. Mycopathologia 179: 111-118.
- Chen YC, Lin YH, Chen KW, Lii J, Teng HJ, et al. (2010) Molecular epidemiology and antifungal susceptibility of Candida parapsilosissensustricto, Candida orthopsilosis, and Candida metapsilosis in Taiwan. Diagn Microbiol Infect Dis 68: 284-292.
- Espinel-Ingroff A (2003) In vitro antifungal activities of anidulafungin and micafungin, licensed agents and the investigational triazoleposaconazole as determined by NCCLS methods for 12,052 fungal isolates: review of the literature. Rev Iberoam Micol 20: 121-136.
- Laverdiere M, Labbé AC, Restieri C , Rotstein C, Heyland D, et al. (2007) Susceptibility patterns of Candida species recovered from Canadian intensive care units. J Crit Care 22: 245-250.
- Hendrickx M, Goffinet JS, Swinne D, Detandt M (2011) Screening of strains of the Candida parapsilosis group of the BCCM/IHEM collection by MALDI-TOF MS. Diagn Microbiol Infect Dis 70: 544-548.
- Quiles-Melero I, García-Rodríguez J, Gómez-López A, Mingorance J (2012) Evaluation of matrix-assisted laser desorption/ionisation time-of-flight (MALDI-TOF) mass spectrometry for identification of Candida parapsilosis, C. orthopsilosis and C. metapsilosis. Eur J Clin Microbiol Infect Dis 31: 67-71.
- Chao QT, Lee TF, Teng SH , Peng LY, Chen PH, et al. (2014) Comparison of the accuracy of two conventional phenotypic methods and two MALDI-TOF MS systems with that of DNA sequencing analysis for correctly identifying clinically encountered yeasts. PLoS One 9: e109376.
Open Access Journals
- Aquaculture & Veterinary Science
- Chemistry & Chemical Sciences
- Clinical Sciences
- Engineering
- General Science
- Genetics & Molecular Biology
- Health Care & Nursing
- Immunology & Microbiology
- Materials Science
- Mathematics & Physics
- Medical Sciences
- Neurology & Psychiatry
- Oncology & Cancer Science
- Pharmaceutical Sciences