Use of Polymerase chain Reaction for Cryptococcus neoformans Genome Detection in Cerebrospinal Fluid for Neurocryptococcosis Diagnosis
Graciele-Melo Cidiane, Rocha-Silva Fabiana, Christo Paulo Pereira and Caligiorne Rachel Basques
DOI10.21767/2471-8521.100013
Graciele-Melo Cidiane1, Rocha-Silva Fabiana1 , Christo Paulo Pereira1,2, and Caligiorne Rachel Basques1*
1Nucleo of Post Graduation and Research of Santa Casa de Belo Horizonte-Hospital, MG, Brazil Rua Domingos Vieira , 59030150240 - Belo Horizonte, Brazil
2Eduardo de Menezes Hospital - Hospital Foundation of Minas Gerais, Brazil
- *Corresponding Author:
- Caligiorne Rachel Basques
Nucleo of Post-Graduation and
Research of Santa Casa de Belo Horizonte-Hospital
MG,Brazil
Rua Domingos Vieira
59030150240 - Belo Horizonte
Brazil
Tel: 55 31 32388677
E-mail: rachelbc@santacasabh.org.br
Received date: April 20, 2016; Accepted date: June 10, 2016; Published date: June 20, 2016
Citation: Graciele-Melo C, Rocha-Silva F, Christo PP and Caligiorne RB. Use of Polymerase chain Reaction for Cryptococcus neoformans Genome Detection in Cerebrospinal Fluid for Neurocryptococcosis Diagnosis. Med mycol Open Access 2016, 2:13. doi: 10.4172/2471-8521.100013
Copyright: © 2016 Graciele-Melo C, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Abstract
Cryptococcosis is a systemic infection caused by encapsulated yeast of Cryptococcus genus, has a worldwide distribution, affecting about 1/3 of human immunodeficiency virus (HIV) carriers and thus, is considered an opportunistic infection. Rapid and specific diagnostic tests for fungal infections are extremely important to effective treatment of infected patients. With the progress of molecular biology has been possible to develop new techniques for diagnosis and Cryptococcus spp identification. This study aimed standardizes a Cryptococcus spp DNA extraction method; determine the detection limit of fungus genome by PCR (Polymerase Chain Reaction) technique and determine the sensitivity and specificity of this technique in biological samples from infected patients. To standardize the DNA extraction were compared two ways for breaking fungus cell wall: the first one, using only glass beads, and the second one, using CTAB reagent. For standardization of conventional PCR and real time PCR (q-PCR), primers previously described were used, specific for Cryptococcus genus (CN4-CN5). A culture of C. neoformans (ATCC 24067) was used as a positive control. For q-PCR standardization for Cryptococcus spp genome detection and quantification in cerebrospinal fluid (CSF) were used 56 samples collected from positive neurocryptococcosis patients, which diagnostic was confirmed by other tests such as India ink, culture, latex and clinical findings. As negative control were used 44 CSF samples collected from negative neurocryptococcosis patients, according the same tests mentioned above. To confirm the specificity of the primers, q-PCR was tested in genomes of other fungal species. According to the results, no significant difference was observed between two DNA extraction methods, suggesting that for DNA extraction in CSF samples it is not necessary to use CTAB to lyse the fungus cell wall. The CTAB residues may inhibit the q-PCR, which is highly sensitive and requires a high degree of purity of the DNA. With respect to the detection limit, it was observed that the lowest DNA concentration detected by q-PCR was 47.0 femtograms (10-5) DNA/µL. The specificity test showed that primers were 100% specific, because it was not aligned in the other fungi species genomes tested. All 56 CSF samples of confirmed neurocryptococcosis patients were positive in q-PCR, while 18 (18.8%) CSF samples among negative neurocryptococcosis patients group were positive in q-PCR. This divergence of results can be explained by the fact that q-PCR has higher sensitivity than other tests used in cryptococcal meningitis diagnosis. Therefore, the q-PCR technique proved to be feasible for neurocryptococcosis diagnosis support, after it is well standardized, as in this study.
Keywords
Neurocryptococcosis; HIV; Cryptococcus spp; Polimerase chain reaction; Molecular diagnosis; q-PCR
Introduction
The neurocryptococcosis is a systemic fungal infection caused by inhaling basidiospores and yeast cells of the fungus Cryptococcus neoformans, which after spending some time in the lungs, is spread through the blood and lodged primarily in the brain and meninges [1,2]. The disease affects about one million people worldwide each year and 400,000 deaths are computed from patients who die within three months after the conception of the disease [3].
The neurocryptococcosis primarily affects patients with immunosuppressive diseases such as acquired immunodeficiency syndrome (AIDS) or lympho proliferative neoplasias, sarcomas, or even those patients that received transplants and are at the use of immunosuppressive therapy [4,5]. The disease emerged in the 80s with the AIDS pandemic, affecting currently, about one-third of individuals carrying the human immunodeficiency virus (HIV), making it the fourth most important cause of death in these patients [6,7].
The fungus tropism for the central nervous system (CNS) is due to the high concentration in the cerebrospinal fluid of important nutrients for fungal growth as thiamine, glutamic acid, glutamine, carbohydrates and minerals [8]. Moreover, the lack of activity of the complement system and weak or absent in brain tissue inflammatory response, enable persistence of fungal cells in the meninges [9,10].
Cryptococcus (Filobasidiella) neoformans is an encapsulated basidiomycete, ubiquitous in the environment and can be found in soil, trees and bird droppings, because its cells are highly resistant to desiccation [11,12]. The species was divided into two varieties: Cryptococcus neoformans var neoformans, with serotypes A, D and AD; Cryptococcus neoformans var gatii, with serotypes B and C [13]. A new variety, Cryptococcus neoformans var grubii, was proposed by Franzot and colleagues [14], with the reclassification of serotype due to molecular and epidemiological differences between serotypes A and D.
The variety neoformans has a worldwide distribution associated with soil contamination by bird droppings and generally is responsible for cryptococcosis in immunocompromised patients, particularly in individuals infected with HIV [11,15-17]. Cryptococcus neoformans var gattii is geographically restricted to the tropical and subtropical climate, commonly found in association with trees species.
Eucalyptos camaldulensis and E. tereticornis, being associated with cryptococcosis in immunocompetent individuals [18-20].
Neurocryptococcosis diagnosis
Laboratory diagnosis of neurocryptococcosis is based on three fundamental methodologies: fresh examination, which consists of yeast observation in clinical material stained with India ink; fungus isolation in artificial culture medium and biochemical tests for identification and search for circulating antigens [13]. Fungus detection is performed in biological samples such as cerebrospinal fluid (CSF), bronchial lavage, secretion of mucocutaneous injuries, urine, macerated tissue obtained by biopsy and bone marrow aspiration [5].
The India ink applied to direct testing of C. neoformans in clinical material provides a differential, faster and cheaper diagnosis, and the analytical sensitivity of the method range from 1000 to 10,000 cells per mL. In cases of HIV patients, which are observed a high burden of yeast, test sensitivity reaches 80% of positive cases of neurocryptococcosis. On the other hand, in immunocompetent patients this percentage can drops to 30-50% [11]. In addiction, this method relies heavily on good technical operator training [21].
Various artificial culture media may be used for C.neoformans growing, such as Agar Sabouraud, Agar niger, Agar L-dopa or dopamine [22]. A positive urine culture may be indicative of disseminated cryptococcosis, even in the absence of clinical signs involving the urinary tract, especially in patients with severe immunodeficiency [23,24]. The cultivation of CSF samples on artificial medium is time consuming, requiring at least 4 days to detect C. neoformans positive cultures and, in some instances, the liquor may result in negative cases due to the small load of viable microorganisms, making it a problem in the monitoring of chronic patients [25].
Serological tests can be used to aid in the diagnosis of cryptococcosis, such as agglutination tests with latex particles, to search polysaccharide antigen of this fungus [26]. ELISA (Enzyme Linked Immunosorbent Assay) for detection of C.neoformans antigens has been described by Casadevall et al. [27] and Mukjerjee & Casadevall [28]. This test has many advantages, including a clear discrimination between positive and negative results, quantitative and highly sensitive information [29]. The biological materials used for testing are cerebrospinal fluid, serum or urine [11].
High antigen securities generally correlate with disease severity and likewise a decrease corresponds to a good prognosis. False-positive reactions may occur related to rheumatoid factor, diseases caused by Trichosporon beigelii and Gram-negative bacilli [30-34].
The molecular methods such as polymerase chain reaction (PCR) represent an excellent alternative for early cryptococcosis diagnosis in comparison with the conventional methods, since it is able to detect lower fungal burden and can be used in small quantities of biological samples without cultivation [35-41].
Ribosomal subunit genes (rDNA) have been exploited for fungal molecular taxonomy and identification, due to the high degree of variability of its nucleotide sequences, which allow distinguishing between genus, species and varieties of fungi [42]. Thus, this study was conducted in order to test the PCR technique application to detect Cryptococcus spp. regions of rDNA in CSF samples and, therefore applied to the diagnosis of cryptococcal meningitis. Once developed, this technique can be used routinely in clinical laboratories, aiding in the diagnosis and cure of disease monitoring.
Materials and Methods
Extraction of genomic DNA
In order to evaluate the efficacy of fungal DNA extraction in cerebrospinal fluid (CSF) samples were compared two different methods of DNA extraction. For both methods, were used 10 CSF samples from positive neurocryptococcosis patients, confirmed by fresh examination stained with India ink and/or by culture findings and/or by Latex test. The patients were examined in Eduardo de Menezes Hospital, Belo Horizonte, Brazil and the CSF samples were kindly provided by the laboratory CSF collection.
The first DNA extraction protocol used glass beads to break the fungus cell wall. For this method, were added 100 mg of sterile glass beads in a sterile 2.0 mL microtube containing 500 μL of Lysis I sterile buffer (3M MW=342.3 Sucrose, 10 mM Tris- HCl pH 7.5, 5 mM MgCl2, Triton X-100 1%) and 500 μL of each CSF sample. The suspension was stirred vigorously on magnetic stirrer (3 pulses of 30 seconds). Was added 10 μL of Proteinase K 20 mg/mL, gently homogenizing the tube and incubating for 30 minutes at 65°C. After incubation, was added to 250 μL of sterile Lysis II buffer (0.075 M NaCl, 0.024 M Na- EDTA pH 8.0 and sterile milli-Q water qs 100 ml) and 5μL of sodium dodecyl sulfate (SDS) at 20%, stirring in vortex for 30 seconds. After that, was added 15 mL of 6 M Sodium Chloride (NaCl), stirring again in vortex for 30 seconds and centrifuged for 15 minutes 5,000 g. The resulting supernatant was recovered into a new sterile 1.5 mL microtube and added to an equal volume of cold absolute isopropanol, homogenized and waited for 15 minutes until the total precipitation of DNA. After precipitation of the DNA, the microtube was centrifuged for 5 minutes at 14,000 g and the supernatant was discarded. The precipitated DNA was washed three times with 1 ml of 75% Ethanol solution. After each wash the microtube was centrifuged at 14,000 g for 1 minute. The final supernatant was discarded, leaving the microtube inverted on paper filter for dry at room temperature until total evaporation of organic residues. After, was added 100 μL of sterile milli-Q water and this was incubated for 10 minutes at 65°C thermo block for hydrating DNA.
The second DNA extraction protocol was used the bromide reagent cetyltrimethylammonium (CTAB) [43], without using glass beads. The DNA extraction using organic solvent CTAB is very applied in plant tissues, biological materials, blood, body tissue, muscle tissues, fungi and bacteria, being an inexpensive method and easy to apply in laboratories [44,45]. In this protocol were used the same procedures as described in the first protocol; however, were not added glass beads in Lysis I sterile buffer and added 700 uL CTAB (20 ml Tris-ClPh 7.5, 20 mL 41% NaCl, 20 ml 0,5M EDTA; 20 mL 10% CTAB and sterile milli-Q water qs 100 mL) to each CSF sample.
Following the procedures, DNA samples was measured by absorbance at 260/280 nm, using the equipment NanoVue Plus–GE (USA).
Conventional PCR
To standardize the PCR reactions a culture of C. neoformans (ATCC 24067) was used as Positive control. The pair of primers CN4 Forward (5'-ATC ACC CTA CCA TTC ACA CATT-3 ') and CN5 Reverse (5'-GAA GGG CAT GCC AGA TGT TTG-3') was chosen [46]. It was used Blastn program (blast.ncbi.hlm.nih.gov/ blast.cgi) to assess the specificity in silico of these primers to Cryptococcus genus.
The PCR reaction was performed in final volume of 15 μL, containing sterile milli-Q water qs, 3.0 mM MgCl2 (Ludwig BIOTEC, Brazil), 0.2 mM dNTPs (Ludwig BIOTEC, Brazil), 0.25 mM of each CN4-CN5 initiator, 1 U Taq DNA polymerase and PCR reaction buffer (Ludwig BIOTEC, Brazil, 47.0 ng/μL DNA template. PCR reactions were performed in Thermal Cycler thermocycler Verite - Applied Biosystems (USA), by means of the initial denaturation at 95°C for 5 minutes, followed by 35 cycles at 95°C for 30 seconds for denaturation, 62°C for 30 seconds for annealing, 72°C for 30 seconds for extension. It also included a final extension cycle at 72°C for seven minutes. The amplifications products were visualized on a 7% polyacrylamide gel, stained by Silver Nitrate 0.2%.
Real-time PCR (q-PCR)
Real time PCR was standardized using the Power Sybr Green Mastermix (Applied Biosystems, Brazil). The reactions were performed in the equipment ECO Real-Time PCR-ILLUMINA (USA).
Standardized q-PCR was performed following the steps recommended in the (MIQE) Johnson et al. [47] as described below:
I- Array of primers was performed by varying the concentration of CN4 and CN5 primers of 0,3 μM, 0,15 μM and 0.25 uM. We used the DNA sample C. neoformans ATCC 24067 (47.0 ng/uL); 1X mastermix Power Sybr Green and sterile ultrapure water (qs 10μL). All concentrations were tested in triplicate. The run was performed as absolute quantification, following the pattern of cycling: 10 minutes at 95°C; 40 cycles of 95°C-10 seconds and 62°C-30 seconds; followed by Melt curve: 95°C-15 seconds, 55°C-15 seconds and 95°C-15 seconds. At the end, the reaction data were analyzed in the equipment program-Eco-software version 4.0. The parameters to determine the best primer concentrations were higher ΔRN by considering the final cycle of amplification in a smaller Cq with a lower standard deviation of the triplicates.
II- q-PCR reaction used 1X Sybr Green Mastermix Power, 0.15 mM each primer and sterile ultrapure water. The cycling conditions were 95°C-10 minutes, 40 cycles of 95°C-10 seconds and 62°C-30 seconds, followed by Melt curve: 95°C-15 seconds, 55°C-15 seconds and 95°C-15 seconds. To confirm the amplification of the expected 136 base pairs fragment, the final reaction product was visualized on a 7% polyacrylamide gel, stained by Silver Nitrate 2%.
Determination of detection limit and PCR efficiency curve
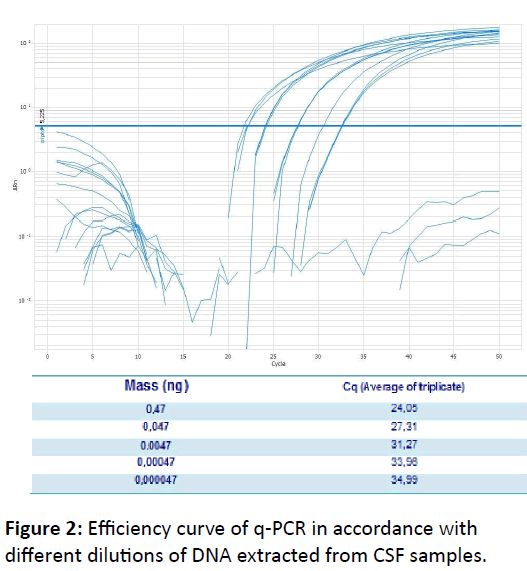
To study the detection limit of PCR were used genomic DNA extracted from C. neoformans ATCC-24067 using Protocol 1, which did not use CTAB. From initial concentration of 47.0 ng/ μL, a serial dilution was made starting at 1:10 in a final volume 50 μL (5 DNA in 45 μL sterile ultrapure water). From the first 1:10 dilution were carried out serial dilutions (10 -2, 10 -3, 10 -4, 10 -5, 10 -6, 10-7 ng/μL). Efficiency curve was analyzed following MIQE parameters, which evaluates the slope, the R2 and efficiency parameters. Ideal slope was between -3.2 and -3.5, R2>0.98 and efficiency of 90 to 105%. The detection limit is the lowest detectable DNA mass.
Specificity in vitro test
To confirm in vitro specificity of primers, the PCR reaction was performed using genomes extracted from another pathogenic fungi: Candida albicans-ATCC 52A and ATCC 18804, Candida krusei- ATCC 6258, Candida glabrata- ATCC Y59, Paracoccidioides brasiliensis- Pb18 and P. lutzii- Pb01. Samples of Aspergillus niger, A. flavus, A. fumigatus, Sporothrix sp. and Histoplasma capsulatum, belong to the collection of Mycology Laboratory of IEP/Santa Casa de Belo Horizonte Hospital. For this reaction primer concentrations and cycling conditions were the same as in previous tests. The analysis was based on samples amplification or not, in any cycle, even in the final. The analysis method was recommended by MIQE in assays for pathogen detection - End Point analysis or presence/absence [47].
Confirming q-PCR amplifications in CSF samples
After standardization, q-PCR was tested in 100 CSF samples, also provided by laboratory CSF collection from Eduardo de Menezes Hospital, Belo Horizonte, Brazil. Among these samples, 56 were from positive neurocryptococcosis patients, confirmed by fresh examination stained with India ink and/or by culture findings and/or by Latex test. The remaining 44 samples were from negative neurocryptococcosis patients, according to above mentioned tests.
Results and Discussion
Comparison of different DNA extraction methods in CSF samples
In the first DNA extraction in CSF samples protocol, which was used glass beads to break the fungi wall, the average of 10 DNA extracted measurements was 11.3 ng/μL and in the second protocol, which was used CTAB to break the fungi wall, the average of 10 DNA extracted measurements was 9.9 ng/ μL. There was no significant difference between extractions methods, being P=0.545 (P>0.05), calculation was performed by Mann Whitney nonparametric method in the program Graphpad Prism 5 (Figure 1).
The results suggest that for DNA extraction in CSF samples it is not necessary to use CTAB to lyse cell wall of the fungus. The CTAB is a detergent commonly used for cell wall lysis, but their residues may inhibit the q-PCR, which is highly sensitive and requires a high degree of purity of DNA [45,48]. As demonstrated by Tanaka et al. [38], glass beads promote mechanical destruction of C. neoformans capsule, being a suitable DNA extraction process and furthermore, not generate chemical waste, being a safer alternative to working with sensitive techniques such as q-PCR.
Primers selection and research in silico and in vitro
The selected pair of primers showed no complementarity with human genome and other pathogenic fungi, according to in silico analysis performed in Blastn software.
Primers were aligned with genomic sequences of the Cryptococcus species. The primers CN4-CN5 flanking ITSrDNA region, generating a fragment of 136 bp, binding in genoma of the three varieties Cryptococcus neoformans var. grubii, Cryptococcus neoformans var gatti and var neoformans (data not show). Despite this primer pair does not differentiate between Cryptococcus varieties, it can be an important tool for rapid diagnosis, enabling the planning of treatment or cure control, already suggested by other studies [49].
According to in vitro results for q-PCR, CN4-CN5 primers showed 100% specificity since there was no amplification of any genome of tested species, demonstrating that it can be used in Cryptococcus genomic DNA detection. These data are confirmed by the research of other authors [35,46,50].
Detection limit of the PCR techniques
According to the results obtained from this study, conventional PCR showed a detection limit of 0.0047 ng/μL, whereas the q-PCR presented a detection limit of 0,000047 ng/μL (Figure 2). Thus, q-PCR detection limit is 100 times less than that of conventional PCR, demonstrating the high sensitivity of q-PCR. This result indicates that q-PCR can be very useful to control cure once fungal load tends to decrease with treatment of the patient and also, can be used in diagnostic in immunocompetent patients, who typically have low fungal load, according describe by Casadevall & Perfect [11].
Moreover, the technique has a high sensitivity, since the 56 samples confirmed positive by routine laboratory tests were all positive in q-PCR (Figure 3 & 4). Among 44 negative samples, 8 (18.18%) were positive on q-PCR. Several factors must be considered when analyzing samples that were positive on q- PCR and negative in routine of Hospital laboratory, among them the low fungal burden in the CSF, which can hamper the growth of fungus in culture or its viewing in fresh examinations [11]. It may also be that DNA is detected from fungal cells which are not viable and remained in liquor after treatment, since these samples were from patients who were under treatment. We have no treatment date about these neurocryptococcosis negative patients.
These results open perspectives for q-PCR use in neurocryptococcosis´s diagnosis. Some studies have demonstrated the q-PCR sensitivity and specificity for genome of fungi detection in biological samples as cerebrospinal fluid, serum and urine [21,36,50]. PCR is an important tool to be used in cryptococcosis diagnosis as it has greater sensitivity than culture, and fresh examination stained with India ink.
In a study conducted by Paschoal et al. [35], PCR was compared with two other Cryptococcus detection techniques, and was shown that although the fresh examination is a faster technique, the sensitivity is only 85.7%. The culture had a sensitivity of 70.8%, while the PCR showed 92.9% sensitivity. The specificity was 100% for all three techniques. In another important study, Leal et al. [49], have standardized a method of multiplex PCR that allows differentiation between C.neoformans var. neoformans and var. Gatti, without samples cultivation, making diagnosis more secure and agile.
In addition, PCR is an improvement on the serology once it detects the pathogen genome in cerebrospinal fluid where it is not possible to detect circulating antibodies, becoming an important tool for diagnosis of infectious diseases on CNS such as neurocryptococcosis.
Rapid and specific diagnostic tests for fungal infections enable more effective treatment and, therefore decrease the morbidity and mortality of diseases [10]. With the advancement of molecular biology has been possible to develop new techniques for diagnosis and serotype identification of Cryptococcus sp, enabling the identification of serotypes from biological materials without necessarily cultivating fungus [50]. In addition to the speed on detection of fungos´s genome, PCR have shown reduced costs of diagnoses each year, making it one of the most cost tools / benefit [21,35,36].
Acknowledgements
The authors thanks to Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq) and Fundação de Amparo à Pesquisa do Estado de Minas Gerais (FAPEMIG) for financial support.
References
- Winkes BL, Mayorga ME, Edman U, Edman JC (1996) Dimorphism and haploid fruiting in Cryptococcus neoformans: association with the ?-mating type. ProcNatlAcadSci 93: 7327-7331.
- Buchanan K, Murphy JW (1998) What makes Cryptococcus neoformans a pathogen?. Emerging Infectious Diseases 4: 71-83.
- Vidal JE, Penalva De Oliveira AC, Dauar RF, Boulware DR (2013) Strategies to reduce mortality and morbidity due to AIDS-related cryptococcal meningitis in Latin America. Braz J Infect Dis 17: 353–362.
- Birchard SJ, Sherding RG (1998) Small Animal Clinic, Saunders Manual, Roca 1406-1408.
- Negroni R (2012) Cryptococcosis. ClinDermatol 30: 599-609.
- Fortes A, Assato M, Fortes S, Lazera M, Wanke B (2002) Meningoencefalitepor Cryptococcus neoformansvariedadegatiiemindígena HIV negativo. American Society for Microbiology 22: 521-528.
- Prado M, Silva MB, Laurenti R, Travassos LR, Taborda CP (2009) Mortality due to systemic mycoses as a primary cause of death or in association with AIDS in Brazil: a review from 1996 to 2006. MemInstOswaldo Cruz 104: 513-521.
- Severo CB, Gazzoni AF, Severo LC (2009) Capitulo 3-Criptococose pulmonar. J bras pneumol 35: 1136-1144.
- Severo LM, Oliveira FM, Silva VB (1998) Differences clinical, epidemiological and ecological between two varieties of Cryptococcus neoformans. Medical Journal Sta Casa Porto Alegre 9: 1672-1686.
- Moretti ML, Resende MR, Lazera MS, Colombo AL, Shikanai-Yasuda MA (2008) Guidelines in cryptococcosis – 2008. Journal of the Brazilian Society of Tropical Medicine 41: 524-544 .
- Casadevall A, Perfect JR (1998) Cryptococcus neoformans. American Society for Microbiology, Washington.
- Pappalardo MC, Melhem MS (2003) Cryptococcosis: a review of the Brazilian experience for the disease. Revista do Instituto de Medicina Tropical de Sao Paulo 45: 299-305.
- Sidrim JJ, Costa AK, Cordeiro RA, Brilhante RS, Moura FE (2010) Molecular methods for the diagnosis and characterization of Cryptococcus: a review. Can J Microbiol 56: 445-58.
- Franzot, Salkin S, Casadevall F (1999) Cryptococcus neoformansvar.grubii: estadovarietal separadopara Cryptococcus neoformanssorotipoAIsolados. J ClinMicrobiol 37: 838-840.
- Fernandes O, Costa T, Costa M (2000) Cryptococcus neoformansisolados de pacientes com AIDS. Journal of the Brazilian Society of Medicine 52: 75-78.
- Ohkusu M, Tangonan N, Takeo K, Kishida E, Ohkubo M, et al. (2002) Serotype, mating type and ploidy of Cryptococcusneoformansstrains isolated from patients in Brazil. Rev Inst Med trop S Paulo 44: 299.
- Soares MCB, Paula CR, Dias ALT, Caseiro MM, Costa SOP (2005) Environmental strains of Cryptococcus neoformans variety grubii in the city of Santos, SP, Brazil. Journal of Tropical Medicine Institute of Sao Paulo 4: 31-36.
- Ellis D, Pfeiffer TJ (1990) Natural habitat of Cryptococcus neoformansvargatii. Journal Clinical Microbiology 28: 1642-1644.
- Licea BA, Garza DG, Urbieta VF, Olivares RAC (1999) Isolation and characterization of Cryptococcus neoformansvar . gatii from samples of Eucalyptus camaldulensisem Mexico City. Iberoamerican Journal of Mycology 16: 40-42.
- Montenegro H, Paula CR (2000) Environmental isolation of Cryptococcus neoformans var. gatii and Cryptococcus neoformans var. neoformans in the city of Sao Paulo. Med Mycol 38: 385-390.
- Rappelli P, Are R, Casu G, Fiori PL, Cppuccinelli P, et al. (1998) Development of Nested PCR for Detection of Cryptococcus neoformans in Cerebrospinal Fluid. J ClinMicrobiol 36: 3438-3440.
- Pedroso RS, Costa KRC, Ferreira JC, Candido RC (2007) Evaluation of melanin production by Cryptococcus species in four different culture media. Rev Soc Bras Med Trop 40: 566-568.
- Calvo B, Fischman O, Castelofilho A, Reis-Filho J, Del Bianco, et al. (1991) Deteccion de antígeno del polisacarido capsular de Cryptococcus neoformansempacientes com SIDA Y neurocriptococosis en São Paulo. Journal of Tropical Medicine Institute of Sao Paulo 33: 485-490.
- Saag MS, Cloud GA, Graybill JR, Sobel JD, Tuazon CU (1999) A comparison of itraconazole versus fluconazole os maintenance therapy for AIDS-associated cryptococcal meningitis. Nacional Institute of Allergy and Infectious Diseases Mycoses Study Group. Clinical Infectious Dieases 28: 291-296.
- Mukjerjee S, Byrne TC, Madden JF, Reller LB (1996) Duration of incubation of fungal cultures. J ClinMicrobiol 34:1583–1585.
- Machado LR, Livramento JA, Vianna LS (2013) Cerebrospinal fluid analysis in infectious diseases of the nervous system: when to ask, what to ask, what toexpect. ArqNeuropsiquiatr 71: 693-698.
- Casadevall A, Mukherjee J, Scharff D (1992) Monoclonal antibody based ELISAs for Cryptococcus polysaccharide. Journal Immunology Methods 154: 27-35.
- Mukjerjee S, Casadevall A (1995) Sensitivy sandwich enzyme-linked immunosorbent assay for Cryptococcus neoformans polysaccharide antigen is dependent on the isotype of the capture and detection antibodies. Journal Clinical Microbiology 33: 765-768.
- Dupont B, Graybill JR, Armstrong D, Laroche R, Touze JE, et al. (1992) Fungal infections in AIDS patients. J Med Vet Mycol 30: 19-28.
- Schoch CL, Seifert KA, Huhndorf S, Robert V, Spouge JL, et al. (2012) Nuclear ribosomal internal transcribed spacer (ITS) region as a universal DNA barcode marker for Fungi. ProcNatlAcadSci 109: 6241-6246.
- Campbell CK, Payne Al, Teall AJ, Brownell A, Mackenzie D (1985) Cryptococcal latex antigen test positive in patient with Trichosporonbeigelli infection. Lancet 2: 43-44.
- BrandaoAfm, Lima As, Rodrigues Abn, LobatoFjc, Santos I (2002) Cryptococcalmeningoencephalitis : clinical study of epidemiological 59.
- Engler HD, Shea YR (1994) Effect of potential interference factors on performance of enzyme immunoassay and latex agglutination assay for cryptococcal antigen. J ClinMicrobiol 32: 2307–2308.
- Hay RJ, Mackenzie DWR (1982) False positive latex tests for cryptococcal antigen in cerebrospinal fluid. J ClinPathol 35:244–245.
- Stamm AM, Polt SS (1980) False-negative cryptococcal antigen test. JAMA 244: 1359.
- Paschoal RC, Hirata MH, Hirata RC, Melhem MS, Dias ALT, et al. (2004) Neurocryptococcosis: diagnosis by PCR method. Rev Inst Med trop S Paulo 46.
- Saha DC, XessBiswas A, Bhowmik DM, Padma MV (2009) Detection of Cryptococcus by conventional, serological and molecular methods. J Med Microbiol 58: 1098-1105.
- Casali AK, Charley CS, Augusto S, Marilene HV (2001) Cryptococcus neoformans-Aspectosmoleculares e epidemiologicos. BiotecnologiaCiencia&Desenvolvimento 20.
- Tanaka K, Miyazaki T, Mitsutake K, Kakeya H, Yamamoto Y, et al. (1996) Detection of Cryptococcus neoformans gene in patients with pulmonary cryptococcosis. J ClinMicrobiol 34: 2826-2828.
- Bialek R, Ibricevic A, Aepinus C, NajvarLk, Fothergill (2000) Detection of Paracoccidioidesbrasiliensis in Tissue Samples by a Nested PCR Assay. J ClinMicrobiol 38: 2940-2942.
- Goldani LZ, Sugar AM (1998) Short report: use of the polymerase chain reaction to detect Paracoccidioidesbrasiliensis in murine paracoccidioidomycosis. J Trop Med Hyg 58: 152-153.
- Lindsley MD, Hurst SF, Iqbal NJ, Morrison CJ (2001) Rapid Identification of Dimorphic and Yeast-Like Fungal Pathogens Using Specific DNA Probes. J ClinMicrobiol 39: 3505-3511.
- Gerrits Van Den Ende A, Hoog GS (1999) Variability and molecular diagnostics of the neurotropic species Cladophialophorabantiana. Studies in Mycology 43: 151-162.
- Moreira M (2010) Methodological variations in the isolation of genomic DNA from Streptococcus bacteria. Braz arch boil technol 53.
- Sollero BP, Faria DA, Paiva SR, Facioni G, Lopes PS (2004) Rapid Method of DNA extraction using CTAB in muscle tissues of pigs.Braz arch boil technol 42: 286-291.
- Mitchell TG, Freedman TJ, White, Taylor JW (1994) Unique oligonucleotide primers in PCR for identification of Cryptococcus neoformans. J ClinMicrobiol 32: 253-255.
- Johnson G, Nolan T, Bustin SA (2013) Real-Time Quantitative PCR, Pathogen Detection and MIQE. In: PCR Detection of Microbial Pathogens. Methods in Molecular Biology 943: 1-14.
- (2010) 7500/7500 Fast Real-Time PCR System Getting Started for Standard Curve Experiments. Applied Biosystems 126.
- Leal AL (2006) Diferenciacao das especies Cryptococcus neoformans e Cryptococcus gattiiutilizando a metodologia de PCR multiplex e determinacao do perfilepidemiologico de pacientes com meningitecriptococica. Dissertacao de mestrado. Universidade Federal do Rio Grande do Sul- Centro de Biotecnologia-Abril de.
- Martins MD, Brighente KB, Matos TA, Vidal JE, Hipolito DD, et al. (2015) Molecular diagnosis of cryptococcal meningitis in cerebrospinal fluid: comparison of primer sets for Cryptococcus neoformans and Cryptococcus gattii species complex. J Infect Dis Braz 19: 62-67.
Open Access Journals
- Aquaculture & Veterinary Science
- Chemistry & Chemical Sciences
- Clinical Sciences
- Engineering
- General Science
- Genetics & Molecular Biology
- Health Care & Nursing
- Immunology & Microbiology
- Materials Science
- Mathematics & Physics
- Medical Sciences
- Neurology & Psychiatry
- Oncology & Cancer Science
- Pharmaceutical Sciences